Genetic alterations
of CDX1, CYLD and CDKN2B genes in CRC
Seyed Mohammad Taghi Hamidian 1, Rezvan Azadi 2,
Pooya Rostami 3, Farnaz Azar Shabe 4, Zeynab Khazaee
Kohparc 4*
1 Babol University of Medical
Sciences, Department of Gastroenterology, Babol, Iran
2 Shahid Beheshti University of
Medical Sciences, Department of Medicine, Tehran, Iran
3 New York University, Londgone
Medical Center, Brooklyn, NY, USA
4 Islamic Azad University of Tonekabon
Branch, Department of Biology, Tonekabon, Iran
*Corresponding Author: Zeynab Khazaee
Kohparc
* Email: zeynab_zhazaee_kohparc@yahoo.com
Abstract
Introduction: Colorectal
cancer (CRC) is the third most frequent type of cancer in the world. In this
explanation, genetic variation is associated in all cancers, particularly CRC,
and modifications of numerous genes, such as CDX1, CYLD, and CDKN2B,
are linked to tumorigenesis in CRC. As a result, this research was conducted in
order to determine changes in the expression of these genes.
Materials
and Methods: Specimens of CRC from 72 individuals with confirmation of
pathology report,were provided and bought from the Bio banks. Real-time PCR was
used to examine the expression of CDX1, CYLD, and CDKN2B
genes in tumoral and non-tumoral tissues. These genes' histological
associations with grading and staging for upregulation and downregulation were
examined.
Result: The expression
of CYLD (P = 0.01) and CDKN2B (P = 0.02) were upregulated
significantly, but the CDX1 (P = 0.03) gene expression was decreased.
Correspondingly, there was no significant association between CDX1
downregulation and CDKN2B upregulation with grade, stage, lymph‐node
metastasis (P= 0.02) and distant metastasis. Moreover, the CYLD
expression was also significantly associated with high grade (P = 0.03), high
stage (P = 0.03), lymph‐node metastasis (P= 0.05) and distant metastasis (P=
0.05).
Conclusion: The
upregulation of CYLD and CDKN2B genes and downregulation of CDX1
gene in tumoral tissues were impressive. Conclusively, the alteration of these
genes expression can be considered as a colorectal cancer biomarker.
Keywords: Colorectal
cancer, CDX1, CYLD, and CDKN2B genes, Alterations
Introduction
Colorectal
cancer (CRC) is one of the most important causes of cancer mortality in the
world (1). The major factor of CRC is the
presence of polyps in the colon and also the changes of adenoma to carcinoma
process. CRC is the growth of cancer cells in the colon part caused by
uncontrolled growth of cells that can proliferate in other tissues irregularly (2). In this way, the term survival of
patients with CRC has not been improved in a therapeutic manner. Strongly,
there is a vital and emergency requirement for a better understanding in the molecular
pathogenesis of CRC in order to recognize the novel biomarkers for prognosis
and diagnosis of CRC (3). Correspondingly, molecular genetic methods especially based on
DNA and RNA investigating are really practical and useful in diagnostic
medicine (4).
CDX1 (caudal-type
homeobox 1) is a transcriptional factor and controls enterocyte differentiation
in the colon, where its expression is different from the crypt-base stem cell
structure. Remarkably, CDX1 is also a keyword to the capacity of a CRC
cell line in differentiation, and it is classified as a negative marker of CRC
stem cells. CDX1 is required for the actual development of the
homeostasis of the intestinal epithelium and also intestinal tract (5). Interestingly, CDX1 is
involved in the modulation of a variety of processes comprising cell adhesion,
columnar morphology, proliferation, and apoptosis. CDX1 is a primary
controller of enterocyte differentiation and its expression is vital for the
transcriptional regulation of a large number of intestine-specific genes
essential for the maintenance of the intestinal phenotype, differentiation, and
intestine development. Many markers in the differentiation process, containing
villin and cytokeratin 20, have been indicated to be directly transcriptionally
regulated by this gene.
Many evidence indicates the loss or down-regulation of CDX1
expression in colon cancer tumors and cell lines (6, 7).
Another
important gene in gastrointestinal cancers particularly CRC, is the cylindromatosis
(CYLD) gene, which was initially explored as a tumor suppressor mutated
for familial cylindromatosis (8). In addition to skin tumors caused
by CYLD loss, decreased CYLD expression has been described in
several types of human cancers comprising breast cancer, hepatocellular
carcinoma, cervical cancer, renal cell carcinoma, lung cancer, gastric cancer
and also colon cancer. Remarkably, the expression profile and clinical
significance of CYLD in patients with a series of co- colorectal lesions
are so important (9-11).
CYLD was recognized
identified as a gene mutated in familial cylindromatosis (FC), a genetic case
that predisposes patients for the progression of skin tumors, termed
cylindroma. Cylindromas are benign tumors that emerge on the scalp and
interestingly is to be derived from hair follicles of stem cells (12). The cylindromatosis patients possess
heterozygous germ-line mutations in the CYLD gene, but the wild-type CYLD
allele undergoes loss of heterozygosity (LOH) and rarely somatic mutations in
different tumors as tumor suppressor gene. The human CYLD gene is
situated on chromosome 16q12.1 and encodes a protein of 956 amino acids. The
C-terminal region of CYLD includes a catalytic domain with sequence
homology to USP family members (9, 13). The second important gene is CDKN2B
which is referred to the CDKN2A tumor suppressor gene in a region at
9p21 and this gene is regularly mutated and omitted in many different tumors.
Considerably, this gene encodes a cyclin-dependent kinase inhibitor, and it is
considered as CDKN2B protein, which is a cell cycle regulator (14). The CDKN2B gene encodes for
CDKN2B, which is a member of the INK4 class of cell cycle
inhibitors. Noticeably, CDKN2B has ankyrin repeats that permit it to
bind and interact of cyclin-dependent kinase (CDK) 4/6 with cyclin D,
through inhibiting the function of CDK4/6. Given the critical role of CDK4/6
and cyclin D in improving development through the G1 checkpoint, CDKN2B
performs as a significant inhibitor of cell cycle and cell proliferation (15, 16).
Materials and Methods
Samples
collection
The research was performed on 72 patients (53 female and 19 male)
which was confirmed by the pathology department and also an agreement by
patients. The histopathological status of patients is shown in Table 2. 72
tumoral and 72 non-tumoral (margins
tissues) were provided and bought from the Bio banks. In this way, DEPC (diethylpyrocarbonate)
was employed to clean and treat all sampling instruments during providing the
biopsies (tumoral and nontumoral tissues) in order to avoid RNAs enzyme.
Correspondingly, after sampling operation, all specimens were transferred to
liquid nitrogen for deep freezing. Vitimately, tissue samples were stored at −
80 °C for long preservation and study. RNA isolation from human tumoral and
nontumoral tissues was performed using a commercial reagent, Trizol (Invitrogen
cat no 15596-025, USA.) Less than 1cm of each tissue was crushed in order to
powder them by a mortar and pestle in the presence of liquid nitrogen, and 40–
80 mg of powdered tissue was used for RNA isolation according to the
manufacture’s protocol. RNA quantity was measured by A260/A280 ratio using
NanoDrop spectrophotometer (TC100, USA) and also controlled by electrophoresis
on agarose gel 2% in order to observe all RNA bands (5S, 18S and 28S).
Relatively, cDNA synthesis was done in the presence of 1 pg total
RNA, 4 μL 5X reaction buffer, 10 mM each of dNTPs, and 1 μL (200 U/ μL) by
QuantiTect Reverse Transcription Kit (cat no 20S313, USA) in a final volume of
20 μL, by 60 min incubation at 44°C. Meanwhile, Real-time PCR was done on
Exicycler q6, Bioneer, USA by using a universal reverse primer and Universal
Taqman-specific probe and also the expression levels of all these genes were
normalized against GAPDH, RNA as control. The 20 μL PCR comprised 1μμL RT
yeild, 0.25 mM universal-specific probe, 0.5 mM each forward and reverse primers.
The PCR reagents were all from Qiagen HotStarTaq reagent set (Qiagen, cat no
203205). The mixtures were incubated at 96 °C for 5 min, followed by 43 cycles
of 90 °C for 45 s, and 63 °C for 1 min. All reactions were done in triplicate.
The CTs were described as the fractional cycle number.
The primers were designed by Allel ID version 7 software. The first
cDNA strand was synthesized. The sequences of forward and reverse primers used
are given in Table 1. The Real-time PCR tests were accomplished in a Step one
instrument (Applied Biosystem, USA) using cDNA. An amount of 1 μl cDNA from
each sample was determined for amplification. GAPDH (glyceraldehyde 3-phosphate
dehydrogenase) was employed as a housekeeping gene. Amplification occurred in a
20 μl final volume by initial incubation at 96 °C for 5 min, followed by 43
cycles of 95 °C for 30 s and 60 °C for 1 min. The range of up-regulation or
down-regulation in each sample was measured using the 2-▲▲ ct
method.
Table 1. Sequences of
primers employed for Real-time PCR action.
|
Primer sequence (5′–3′)
|
|
Forward CDX1
|
5´-AAGCCTCCGRRCCGCGAATCA-3´
|
|
Reverse CDX1
|
5'-GGAAGACTCGTGTATGTATGTGY ATATGTG-3'
|
|
Forward CYLD
|
5'-ATGGATAACCCTATTGGCAACTG-3'
|
|
Reverse CYLD
|
5'-GTATCCAGTGCTGCGACCGT-3'
|
|
Forward CDKN2B
|
5'-
TGGCCGGAGGTCATGATG -3'
|
|
Reverse CDKN2B
|
5'- GGGCAGCATCATGCACCG -3'
|
Statistical
Analyses
All the acquired data from Real-time PCR were analyzed by exercycle
set. Correspondingly, the significant difference was statistically interpreted
by paired Student’s t-test. P < 0.05 was considered statistically
significant. Analyses were accomplished using commercially available
statistical software (SPSS Statistics software, version 25, Chicago).
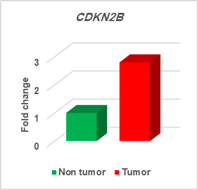
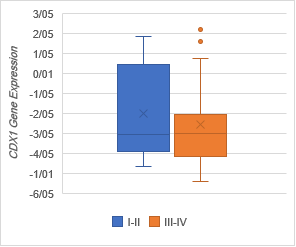
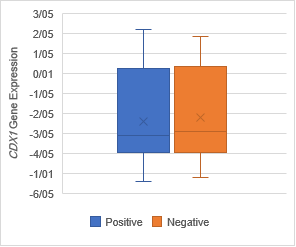
Results
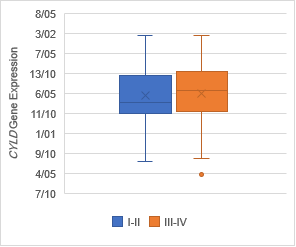
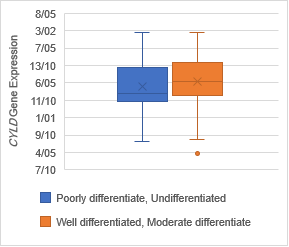
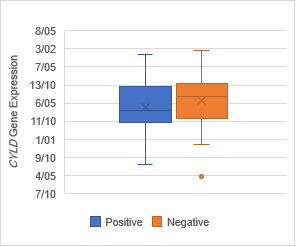
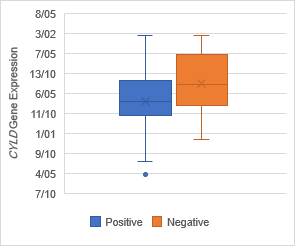
Gene expression
evaluation in tumoral tissues
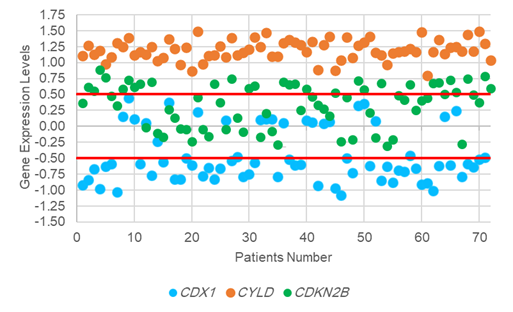
The analysis of expression levels of tumoral and corresponding
non-tumoral tissues for CDX1, CYLD and CDKN2B genes
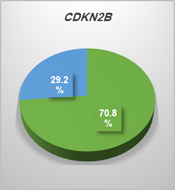
indicated that the CYLD and CDKN2B
were down regulated in tumoral tissues in comparison with their non-tumoral
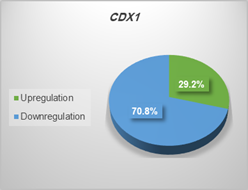
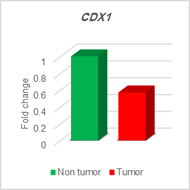
counterparts (P = 0.02). On the contrary, CDX1 expression level had
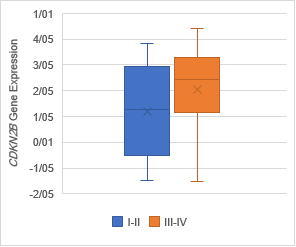
decreased significantly in 70% of samples (Figure 1,2,3).
Discussion
Transgenic
expression of CDX1 in mouse gastric epithelium causes intestinal
transdifferentiation, which protects this consideration that CDX1 is
up-regulated in Barrett’s metaplasia of the esophagus. Considerably, many
transcriptional targets and effective activities of CDX1 have been
recognized, there remains much to learn about the mechanisms by which it
encourages differentiation and, also, those by which it inhibits stemness CDX1
action as transcription factors regulate a wide range of cellular mechanisms (6).
Additionally, CDX1, an intestine-specific transcription
factor, is a candidate tumor suppressor gene and it manages the
intestine-specific gene transcription and regulates the intestinal epithelial
cell phenotype. Past investigation illustrated that the murine CDX1
overexpression in rat normal intestinal epithelial cells regulates
proliferation as a conclusion of inducing cell cycle arrest. Meaningly, this
antiproliferative role may be mediated through down-regulation of the D-type
cyclins (17). The CDX1
gene is expressed in a collaborative model during intestinal progression. CDX1
expression will last in the intestinal epithelium throughout life, notably in
the crypt. The same model of CDX1 expression was discovered in the human
small intestine. Many searches have described that the CDX1 expression
is markedly down-regulated in both adenomas and carcinomas of the colon. Little
is known about the molecular mechanisms that regulate the developmental and
spatial patterns of the CDX1 expression in normal intestine or what
induces the down-regulation in colonic adenomas and cancers (18). Wong et al.
have shown that the loss or reduction of CDX1 is often induced by
promoter methylation. Together, these observations indicate a potential role of
CDX1 loss in tumor development (19).
Recently, the expression monitoring of CYLD in many
colorectal-related lesions and the clinical significance of CYLD
expression in CRC have remained unclear, although, past investigation
indicating that both the transcription function and the protein level of CYLD
were downregulated in colon cancer in comparison with normal colon tissues. The
difference of CYLD expression in the normal colorectal epithelium,
benign adenoma, primary CRC and metastatic lesions was explored (20). Of particular
interest, we wondered whether CYLD expression played a part in tumor
development, progression, or metastasis and whether reduced CYLD
expression was a good or poor prognostic factor for CRC patients. These
findings strengthened the fact that CYLD functioned as a
tumor-suppressor gene not only in the skin tumor but also in CRC. In addition,
reduced CYLD expression was an independent factor for poor prognosis of
CRC patients. Based on the evidence above, our results also recommended that
the downregulation of CYLD might be involved in a series of important
biological properties of colorectal cancer cells, such as carcinogenesis, tumor
progression and metastasis (21). These
findings also have implications on the tumor suppressor function of CYLD,
as colonic inflammation in IBD patients is a risk factor for colorectal cancer.
The potential association of CYLD gene suppression with colon cancer is
more directly suggested by a study showing reduced expression of CYLD in
colon cancer cell lines and tissue samples It is currently unknown how the CYLD
gene is suppressed in IBD and colon cancer cells. Nevertheless, the mechanistic
insight of CYLD gene repression has been provided by studies using other
cancer models (22).
In another study, CYLD expression was analyzed in two of the
most common human carcinomas worldwide. Colon carcinoma derives from intestinal
epithelial cells and HCC derives from hepatocytes. We found reduced CYLD
mRNA expression in all three HCC cell lines and eight colon carcinoma cell
lines examined compared with normal primary cells. Additionally, reduction or
loss of CYLD expression was found in situ in most hepatocellular and
colon carcinoma compared with non-neoplastic tissue samples. Analysis on
protein level confirmed these findings. Functional assays with CYLD
transfected cell lines revealed that CYLD expression decreased NF-κB
activity. Thus, functional relevant loss of CYLD expression may
contribute to tumor development and progression, and may provide a new target
for therapeutic strategies (11). CDKN2B
is a cyclin-dependent kinase inhibitor and functions as a cell growth regulator
that controls cell cycle G1 progression. Last investigations have acknowledged CDKN2B
as a required tumor suppressor, and deletion of its enhancer element is related
to many different malignancies. Silencing of CDKN2B gene expression by
epigenetic modification characterize in multiple myelomas gastric
adenocarcinoma (23). Reexpression
of CDKN2B in tumor-derived cells significantly attenuates the
tumorigenic potential of the cells and delays tumor progression (24). Fluctuation of CDKN2B's expression has
been announced in association with many malignancies particularly, prostate,
colorectal, breast, and liver cancer. Considerably, CDKN2B were
ubiquitously expressed in colon cancer at different stages of tumorigenesis (25).
CDKN2B encoded by the
INK4b-ARF-INK4a locus. It is an acknowledged tumor suppressor gene that can
form a complex with CDK4 or CDK6 and inhibits the activation of
the cyclin-dependent kinase and progression of the cell cycle. The
INK4b-ARF-INK4a locus is organized by Polycomb repressive complexes. In this
way, downregulation of CDKN2B was investigated in cancers (26). The
epigenetic investigation of these genes alongside gene expression and also a mutation
of other genes which are involved in GI cancers is recommended strongly.
Conclusion
It is concluded that the upregulation of CYLD
and CDKN2B genes and downregulation of CDX1 gene in tumoral
tissues were impressive. Conspicuously, the modification of these genes
expression can be accepted as the main biomarker in colorectal cancer.
Author
contributions
RZ, PR, and FAS
collected data and accomplished some sections of the study and manuscript, SMTH
collected all the biopsies directly in Omid clinic and hospital by himself and
also confirmed the clinical qualifications of all the patients as a gastroenterologist.
ZKK controlled and confirmed the data quality, evaluated and optimized the
informatics database, wrote the paper and edited it, some other essential
functions containing study design, controlling the project and protocol development
and also data analysis. All authors revised the article carefully, read
and acknowledged the final version of the paper.
Acknowledgments
We thank all
people who were involved in this project and contributed us.
Conflict of
interests
Authors declare no conflict of interest.
References
1. Gandomani
HS, Yousefi SM, Aghajani M, Mohammadian-Hafshejani A, Tarazoj AA, Pouyesh V, et
al. Colorectal cancer in the world: incidence, mortality and risk factors.
Biomedical Research and Therapy. 2017;4(10):1656-75.
2. Keum N, Giovannucci E. Global
burden of colorectal cancer: emerging trends, risk factors and prevention
strategies. Nature reviews Gastroenterology & hepatology.
2019;16(12):713-32.
3. Goel G. Molecular
characterization and biomarker identification in colorectal cancer: Toward
realization of the precision medicine dream. Cancer management and research.
2018;10: 5895–5908.
4. Katsanis SH, Katsanis N.
Molecular genetic testing and the future of clinical genomics. Nature Reviews
Genetics. 2013;14(6):415-26.
5. Lynch J, Keller M, Guo R-J,
Yang D, Traber P. Cdx1 inhibits the proliferation of human colon cancer cells
by reducing cyclin D1 gene expression. Oncogene. 2003;22(41):6395-407.
6. Jones MF, Hara T, Francis P,
Li XL, Bilke S, Zhu Y, et al. The CDX1–microRNA-215 axis regulates colorectal
cancer stem cell differentiation. Proceedings of the National Academy of
Sciences. 2015;112(13):E1550-E8.
7. Coskun M, Troelsen JT, Nielsen
OH. The role of CDX2 in intestinal homeostasis and inflammation. Biochimica et biophysica
acta (BBA)-Molecular basis of disease. 2011;1812(3):283-9.
8. Rajan N, Burn J, Langtry J,
Sieber‐Blum M, Lord CJ, Ashworth A. Transition from cylindroma to spiradenoma
in CYLD‐defective tumours is associated with reduced DKK2 expression. The Journal
of pathology. 2011;224(3):309-21.
9. Verhoeft KR, Ngan HL, Lui VWY.
The cylindromatosis (CYLD) gene and head and neck tumorigenesis. Cancers of the
head & neck. 2016;1(1):10.
10. Hayashi M, Jono H, Shinriki S,
Nakamura T, Guo J, Sueta A, et al. Clinical significance of CYLD downregulation
in breast cancer. Breast cancer research and treatment. 2014;143(3):447-57.
11. Hellerbrand C, Bumes E,
Bataille F, Dietmaier W, Massoumi R, Bosserhoff AK. Reduced expression of CYLD
in human colon and hepatocellular carcinomas. Carcinogenesis. 2007;28(1):21-7.
12. van den Ouweland AM, Elfferich
P, Lamping R, van de Graaf R, van Veghel-Plandsoen MM, Franken S, et al.
Identification of a large rearrangement in CYLD as a cause of familial
cylindromatosis. Familial cancer. 2011;10(1):127-32.
13. Aguilera CA, De la Varga
Martínez R, García LO, Jimenez-Gallo D, Planelles CA, Barrios ML. Heterozygous
cylindromatosis gene mutation c. 1628_1629delCT in a family with brook-spiegler
syndrome. Indian journal of dermatology. 2016;61(5):580.
14. Kryh H, Carén H, Erichsen J,
Sjöberg R-M, Abrahamsson J, Kogner P, et al. Comprehensive SNP array study of
frequently used neuroblastoma cell lines; copy neutral loss of heterozygosity
is common in the cell lines but uncommon in primary tumors. BMC genomics.
2011;12(1):443.
15. Scruggs AM, Koh HB, Tripathi P,
Leeper NJ, White ES, Huang SK. Loss of CDKN2B promotes fibrosis via increased
fibroblast differentiation rather than proliferation. American journal of
respiratory cell and molecular biology. 2018;59(2):200-14.
16. Otto T, Sicinski P. Cell cycle
proteins as promising targets in cancer therapy. Nature Reviews Cancer.
2017;17(2):93.
17. Fujii Y, Yoshihashi K, Suzuki
H, Tsutsumi S, Mutoh H, Maeda S, et al. CDX1 confers intestinal phenotype on
gastric epithelial cells via induction of stemness-associated reprogramming
factors SALL4 and KLF5. Proceedings of the National Academy of Sciences.
2012;109(50):20584-9.
18. Suh ER, Ha CS, Rankin EB,
Toyota M, Traber PG. DNA methylation down-regulates CDX1 gene expression in
colorectal cancer cell lines. Journal of Biological Chemistry.
2002;277(39):35795-800.
19. Li X, Wang X, Tan Z, Chen S,
Guan F. Role of glycans in cancer cells undergoing epithelial–mesenchymal
transition. Frontiers in oncology. 2016;6:33.
20. Wang Y, Li Y, Zhou B, Zhang W,
Guan J, Wang R, et al. Expression of the apoptosis inhibitor livin in
colorectal adenoma-carcinoma sequence: correlations with pathology and outcome.
Tumor Biology. 2014;35(12):11791-8.
21. Tauriello DV, Haegebarth A,
Kuper I, Edelmann MJ, Henraat M, Canninga-van Dijk MR, et al. Loss of the tumor
suppressor CYLD enhances Wnt/β-catenin signaling through K63-linked
ubiquitination of Dvl. Molecular cell. 2010;37(5):607-19.
22. Zhao H, Xu F, Jin M, An G, Feng
G. CYLD expression in benign, malignant and metastatic lesions of colorectal
epithelium and its prognostic role in colorectal carcinoma. Int J Clin Exp
Pathol. 2016;9(4):4909-16.
23. Fu D-G. Epigenetic alterations
in gastric cancer. Molecular medicine reports. 2015;12(3):3223-30.
24. Weng X, Zeng L, Yan F, He M, Wu
X, Zheng D. Cyclin-dependent kinase inhibitor 2B gene is associated with the
sensitivity of hepatoma cells to Sorafenib. OncoTargets and therapy.
2019;12:5025.
25. Zhu H, Ji Y, Li W, Wu M.
Identification of key pathways and genes in colorectal cancer to predict the
prognosis based on mRNA interaction network. Oncology Letters.
2019;18(4):3778-86.
26. Xu M, Chen X, Lin K, Zeng K,
Liu X, Pan B, et al. The long noncoding RNA SNHG1 regulates colorectal cancer
cell growth through interactions with EZH2 and miR-154-5p. Molecular cancer.
2018;17(1):1-16.