Regulating and
changeable performance of CDX2, CTNNBIP1, and FAT4 genes
in colorectal cancer
Seyed Mohammad Taghi Hamidian 1, Rezvan Azadi 2,
Pooya Rostami 3, Farnaz Azar Shabe 4, Zeynab Khazaee
Kohparc 4*
1 Babol University of Medical
Sciences, Department of Gastroenterology, Babol, Iran
2 Shahid Beheshti University of
Medical Sciences, Department of Medicine, Tehran, Iran
3 New York University, Londgone
Medical Center, Brooklyn, NY, USA
4 Islamic Azad University of Tonekabon
Branch, Department of Biology, Tonekabon, Iran
*Corresponding
Author: Zeynab Khazaee Kohparc
* Email: zeynab_zhazaee_kohparc@yahoo.com
Abstract
Introduction: Colorectal
cancer (CRC) is the third most frequent type of cancer in the world. In this
explanation, genetic variation is associated in all cancers, particularly CRC,
and modifications of numerous genes, such as CDX2, CTNNBIP1, and FAT4,
are linked to tumorgenesis in CRC. As a result, this research was conducted in
order to determine changes in the expression of these genes.
Materials
and Methods: After obtaining patient consent and pathology department approval,
from72 individuals with confirmation of pathology report,were provided and
bought from the Bio banks. Real-time PCR was used to examine the expression of CDX2,
CTNNBIP1, and FAT4 genes in tumoral and non-tumoral tissues.
These genes' histological associations with grading and staging for
upregulation and downregulation were examined.
Result: CDX2 (P
= 0.01) and CTNNBIP1 (P = 0.03) expression were highly increased,
whereas FAT4 (P= 0.05) expression was downregulated. Similarly, there
was no evidence of a link between CDX2 and CTNNBIP1
overexpression and grade, stage, lymphnode metastasis, or distant metastasis.
Furthermore, FAT4 expression was linked to highe stage, high grade, distant metastasis
and lymphnode metastasis (P 0.05).
Conclusion: CTNNBIP1
and CDX2 genes were upregulated in tumoral tissues, while FAT4
genes were downregulated. Finally, changes in the expression of these genes can
be used as a CRC biomarker.
Keywords: Colorectal
cancer, Genes fluctuation, Regulation
Introduction
Colorectal cancer ( CRC) is one of the most commonly
diagnosed cancer in adults. The third
prevalent cancer in the world is CRC
(1). CRC is a
prevalent human cancer that requires a thorough knowledge of its molecular
underpinnings. Initial therapy only cures a small percentage of people and is
most effective when the disease is in its initial stages (2). CRC was among the first large epithelial
malignancies in which molecular changes were observed systematically as the
disease progressed. The discovery of new oncogenes and tumor suppressors would
help us identify the biology of CRC and could lead to new effective treatments (3).
Since
CDX2 mutations are extremely rare events in CRCs, we hypothesized that
epigenetic changes, such as promoter hypermethylation or histone deacetylation
could be responsible for significant downregulation or absence of CDX2,
particularly in the group of tumors displaying “serrated” molecular features. Human
serrated adenomas with high-grade dysplasia have been shown to have
significantly greater frequencies of CDX2 hypermethylation than other
polyp types (4). CTNNBIP1 (β‐catenin
interacting protein 1) gene is an antagonist of Wnt signaling which binds to
the β‐catenin molecules. The CTNNBIP1 function as a tumor suppressor
gene or oncogene in different types of cancer is controversial. Several nuclear
antagonists are known to regulate β-catenin-TCF mediated transcription. One
such direct nuclear antagonist is CTNNBIP1 (catenin, beta interacting
protein 1; also known as ICAT) (5). CTNNBIP1 binds to two
different armadillo regions of β-catenin through its N-terminal and C-terminal
domains leading to disruption of β-catenin-TCF interaction. The importance of CTNNBIP1
in embryonic development and tissue differentiation process has been reported.
Variable frequencies of expression of CTNNBIP1 have been shown in
metastatic and nonmetastatic human melanoma (6). The Fat gene family was originally
identified in Drosophila as a member of the cadherin super-family with tumor
suppressor functions. It regulates cell proliferation and planar cell polarity
during Drosophila development by the Hippo signaling pathway. They encode a
type 1 transmembrane protein with 34 cadherin repeats, 4 epidermal growth
factor (EGF)-like repeats, a transmembrane domain and a cytoplasmic domain that
is distinct from the classical cadherin proteins. In humans, four members of
the Fat family have been identified, namely, FAT1, FAT2, FAT3 and FAT4,
which are structurally similar to the Drosophila Fat protein. In mammals, FAT4
is the true structural ortholog of the Drosophila FAT. FAT4 functions as
a tumor suppressor and previous findings have demonstrated that FAT4 can
inhibit the epithelial-to-mesenchymal transition (EMT) and the proliferation of
gastric cancer cells. However, few studies have investigated the role of FAT4
in the development of colorectal cancer (7).
Materials and Methods
Samples collection
The
study sample consisted of 72 tumoral and 72
non-tumoral (margins tissues) from 53 females and 19 males were provided
and bought from the Bio banks. Information on histological status is shown in
Table 1. Then, all tissues were delivered to liquid nitrogen for deep freezing.
Tissue samples were kept at a temperature of 80 °C for long-term conservation
and investigation. Trizol (Invitrogen cat no 15596-025, USA.) was used to
isolate RNA from tissues. The spectrophotometer (TC100, USA) was used for quantitative
RNA analysis and electrophoresis (2% agarose gel) was used for qualitative
analysis.
cDNA
was prepared using the cDNA Kit (Quanti Test Reverse transcription kit, Qiagen)
with around2 pg RNA per reaction. The first cDNA strand was generated utilizing
a stem-loop sequence-specific primer. Table 2 lists the forward and reverse
primer sequences. The real-time PCR assays were carried out on cDNA by using the
SYBR Green technique in Step one equipment (Applied Biosystem, USA). A total of
1 liter of cDNA from each tissue was used for amplification. As a housekeeping
gene, GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was employed. Early
incubation at 95 °C for 5 minutes was proceeded by 40 cycles of 95 °C for 30 s
and 60 °C for 1 min in a 20 l final volume. Using the 2-ct approach, the range
of up-regulation or down-regulation in each sample was extensively studied. All
of the reactions were carried out in triplicate.
Table 1. Sequences of primers employed for
Real-time PCR action.
|
Primer sequence (5′–3′)
|
|
Forward CDX2
|
5´-TAGTTTGYGGGGYTGYTGTA-3´
|
|
Reverse CDX2
|
5´-GCCATATACRTAARCTACCTCCT-3'
|
|
Forward CTNNBIP1
|
5′‐GGAAGATGGGATCAAACCTGA CAG‐3′
|
|
Reverse CTNNBIP1
|
5′‐TCGTATCCAGTGCTGCGACCGTAT
GGATGTGTCTGCGGCGTTTTATCATGCACTGGATACGAC AAC GCCATCA CC‐3′
|
|
Forward FAT4
|
5'-ACACTGTGATTGCCAGGAGAG-3'
|
|
Reverse FAT4
|
5'-GGATGTGTCTGCGGCGTTTTAT
CATGCACTGGATACGACCAAGAGTCCAGTC-3'
|
Statistical Analyses
All
the acquired data from Real-time PCR were analyzed by exercycle set.
Correspondingly, the significant difference was statistically interpreted by
paired Student’s t-test. P < 0.05 was considered statistically significant.
Analyses were accomplished using commercially available statistical software
(SPSS Statistics software, version 25, Chicago).
Results
Gene expression evaluation in
tumoral tissues
The analysis of expression levels of tumoral and corresponding
non-tumoral tissues for CDX2, CTNNBIP1 and FAT4 genes indicated
that the CDX2 and CTNNBIP1 were
upregulated in tumoral tissues in comparison with their non-tumoral
counterparts. On the contrary, FAT4 expression level had decreased
significantly in 50% of samples (Figure 1, 2,3).
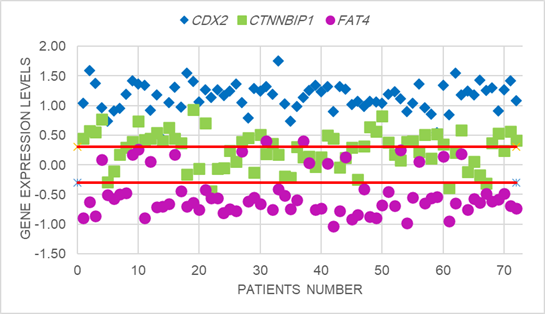
Figure1. Scatter plot analysis of relative
expression of CDX2, CTNNBIP1 and FAT4 in colorectal cancer
patients. The Y-axis indicates the logarithm of relative gene expression.
Horizontal red lines represent cut-off values logarithms for two-fold changes
in expression (FC≥2.0, p<0.05). The upper part of the graphs indicates
up-regulation in the tumoral compared to the non-tumoral tissue; the lower part
of the graph indicates down-regulation in the tumoral compared to the
non-tumoral tissue (differences in expression ≥ 2; P < 0.05). The CDX2
(P = 0.01) and CTNNBIP1 (P =
0.03) expression level had increased and FAT4 (P
= 0.05) expression level had decreased significantly in tumoral compared to
the non-tumoral samples.
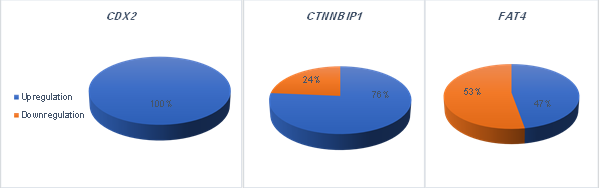
Figure
2. The data
revealed a significant upregulation of CDX2 and CTNNBIP1 expression and downregulation
of FAT4 in colorectal cancer (P < 0.05.
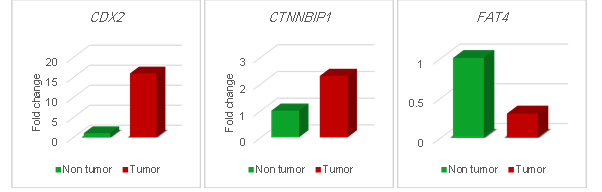
(a)
(b)
(c)
Figure 3. Fold change of (a) CDX2
(P= 0.02), (b) CTNNBIP1 (P= 0.02) and (c) FAT4 (P=
0.04) expression in tumoral tissues in comparison with non-tumoral (tumor
margin) tissues.
Clinicopathological analysis
Clinicopathological
consequences of CDX2, CTNNBIP1 and FAT4 genes expression were
evaluated in 72 patients diagnosed with adenocarcinoma of the colorectal.
Patients’ clinicopathological characteristics are summarized in Table 2. The
analysis of different clinicopathological variables and genes expression
correlation is presented in Table 3 (up/down).
The mean age of patients was 58.9±12.5 years at the time of diagnosis
(female to male ratio, 4:1; age range, 37–88 years). In general, more than half
of the patients had advanced T‐stage (Stages III-IV), and high‐grade histology.
Lymph‐node metastasis and distant metastasis were observed in more than 60% of
the patients.
The number of gene expressions of
all samples was compared and investigated with the stage, grade, lymph node
metastasis and distance metastasis of all patients.
The analysis of different
clinicopathological variables and genes expression correlation is presented in
Table 3. Statistical analyzes were performed with using SPSS 25 and also Chi
Square test and T test.
The expression of CDX2, CTNNBIP1 and
FAT4 was matched with different clinicopathological data of the colorectal
cancer patients (summarized in Table 2). There was no significant association
between CDX2 and CTNNBIP1 expression with grade, stage,
lymph‐node metastasis (P= 0.02) and distant metastasis. Moreover, the FAT4
expression was also significantly associated with high grade (P = 0.03), high
stage (P = 0.03), lymph‐node metastasis (P= 0.05) and distant metastasis (P=
0.05) (Figure 4, 5, 6).
Table 2. Clinicopathological characteristics of colorectal cancer cases.
|
Total (N=72)
Patients (%)
|
Characteristics
|
|
53 (73.6)
19 (26.4)
|
Gender
Female
Male
|
|
38 (52.8)
34 (47.2)
|
Age
< 60 years
≥ 60 years
|
|
6 (8.3)
24 (33.3)
38 (52.8)
4 (5.6)
|
Stage
I
II
III
IV
|
|
4 (5.6)
26 (36.1)
39 (54.1)
3 (4.2)
|
Grade
Well-differentiated
Moderate differentiate
Poorly differentiate
Undifferentiated
|
|
45 (62.5)
27 (37.5)
|
LM
Yes
No
|
|
44 (61.1)
28 (38.9)
|
DM
Yes
No
|
Table 3. The association of genes expression with clinicopathological
qualification. LM: Lymph node Metastasis, DM: Distance Metastasis; ↓/−: decrease
or no change of expression; ↑: increase of gene expression.
|
|
CDX2
|
P-value
|
CTNNBIP1
|
P-value
|
FAT4
|
P-value
|
|
Tumor Stage
I-II
III-IV
|
↓/−
0
0
|
↑
30
42
|
0.5
|
↓/−
12
5
|
↑
18
37
|
0.7
|
↓/−
25
32
|
↑
5
10
|
0.03
|
|
Tumor Grade
I-II
III-IV
|
0
0
|
30
42
|
0.6
|
13
6
|
17
36
|
0.1
|
23
35
|
7
7
|
0.03
|
|
LM
Yes
No
|
0
0
|
44
28
|
0.3
|
24
11
|
22
15
|
0.4
|
36
20
|
8
8
|
0.05
|
|
DM
Yes
No
|
0
0
|
44
28
|
0.2
|
21
15
|
23
13
|
0.5
|
36
21
|
8
7
|
0.05
|
|
|
|
|
|
|
|
|
|
|
|
LM: Lymph node Metastasis, DM: Distance Metastasis
The Association of CDX2, CTNNBIP1 and FAT4 expression
with clinicopathological qualifications
(a)
(b)

(c)
(d)
Figure 4. The Association of CDX2 expression
with clinicopathological qualifications. There was no significant association
between CDX2 upregulation with (a) tumor stage (P =0.5), (b)
tumor grade (P =0.6), (c) lymph‐node metastasis (P= 0.3) and (d)
distance metastasis (P= 0.2).

(a)
(b)

(c)
(d)
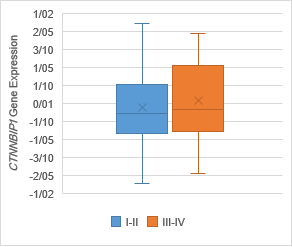
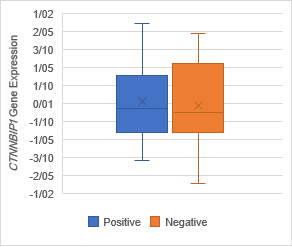
Figure 5. The Association of CTNNBIP1 expression with clinicopathological
qualifications. There was no significant association between CTNNBIP1 downregulation
with (a) tumor stage (P =0.7), (b) tumor grade (P =0.1), (c)
lymph‐node metastasis (P= 0.4) and (d) distance metastasis (P= 0.5).
(a)
(b)
(c)
(d)
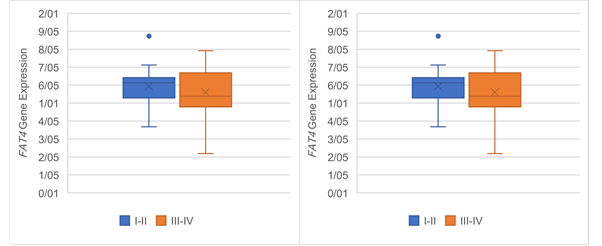
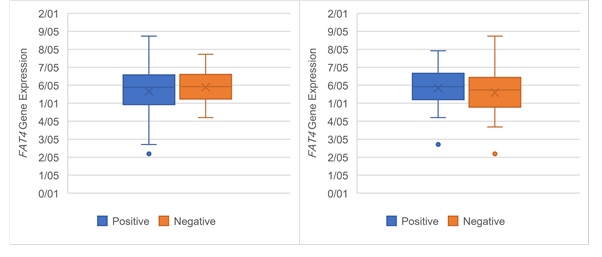
Figure 6. The Association of FAT4 expression
with clinicopathological qualifications. The FAT4 expression was
significantly associated with (a) tumor stage (P =0.03), (b)
tumor grade (P =0.03), (c) lymph‐node metastasis (P= 0.05) and (d)
distance metastasis (P= 0.05).
Discussion
Reduced CDX2 protein expression is related to certain
molecular alterations during colorectal tumorigenesis. Previous work shows that
nearly all sporadic microsatellite unstable (MSI) cancers show some degree of
loss of the protein in the tumor, whether in a small or substantial percentage
of cells. This loss is not however limited to MSI-high cancers but is also
found in microsatellite stable (MSS) tumors with BRAF mutation and high-level
CpG island methylator phenotype (CIMP), in other words, in cancers deriving
from the so-called serrated pathway (4). The previous research showed CDX2
expression was increased significantly in gastric cancer. CDX2
expression had a significant correlation with TNM stage and lymph node
metastasis.
Previous findings have shown that transfection of CDX2 cDNA,
and human HT29 CRC cell line to express CDX2 protein, indicated the
oncogenic potential of the abovementioned cells, and metastasis of related
cells markedly decreased while cell sensitivity for apoptosis significantly
increased. The results have shown that in comparison to the normal population,
the degree of methylation of the promoter region of CDX2 in lesion
tissue of patients with CRC was higher than that of the normal population. The
protein expression in the control and lesion sections of CRC patients showed
that the expression level of CDX2 in the lesion section of patients with
CRC was lower. This finding suggested that there was a certain correlation
between CDX2 and CRC or the decrease in the degree of CDX2 gene
promoter methylation to a certain extent, promotes the risk of CRC (8).
Previous research indicates the downregulation of CTNNBIP1
gene which corresponds to a tumor suppressor role for CTNNBIP1 in GC.
Also, the expression level of CTNNBIP1 was extremely lower in female
patients than males. According to our findings, the tumor-suppressing function
of CTNNBIP1 in GC is mostly associated with initiation procedures,
because well‐differentiated tumors showed significant downregulation of CTNNBIP1
compared with other malignant grades. CTNNBIP1 expression associated
with EBV and CMV infections suggests that the Wnt/β‐catenin dysregulation is
affected by these agents in GC.
CTNNBIP1 is a suppressor of lung cancer progression. The CTNNBIP1
protein is important, in that it can control lung cancer cell migration via the
coordinated regulation of the β-catenin pathway. A low expression of CTNNBIP1
is correlated with a high level of expression of MMP7, and there is also an
upward trend in terms of the pathological stage and poorer patient survival,
which suggests that CTNNBIP1 may be able to serve as a prognostic
biomarker for lung cancer (9).
FAT4 is a tumor suppressor in CRC. Moreover, FAT4 silencing
inhibits CRC cell autophagy and stimulates the invasion and migration of these
cells as well as the EMT, whereas the overexpression of FAT4 yields the
opposite results and increases autophagy. Furthermore, the stimulatory effects
of FAT4 on autophagy occur through the upregulation of LC3 and the
downregulation of P62 and the effects of FAT4 on the EMT, as evidenced
by the detected changes in the expression levels of E-cadherin and Twist1.
Moreover, an increase in FAT4 leads to a reduction in xenograft tumor
growth in vivo, whereas the opposite outcome was obtained with FAT4
knockdown. Therefore, we conclude that FAT4 regulates the activity of
PI3K to promote autophagy and inhibit the EMT, and these effects are partly
achieved through the PI3K/AKT/mTOR and PI3K/AKT/GSK-3β signaling pathways. We
anticipate that this study will provide a basis for establishing new strategic
approaches for the development of effective CRC therapies (10).
Cai et al, found that FAT4
has a tumor suppressor role mediated by the modulation of Wnt/β-catenin
signaling, providing potential novel targets for the treatment of gastric
cancer (11).
Conclusion
The overexpression of CDX2 and CTNNBIP1
expression in tumoral tissues, as well as the downregulation of FAT4,
were found to be outstanding. Interestingly, changes in the expression of these
genes can be used as a primary biomarker in CRC.
Author
contributions
RZ, PR, and FAS collected data and accomplished
some sections of the study and manuscript, SMTH collected all the biopsies
directly in Omid clinic and hospital by himself and also confirmed the clinical
qualifications of all the patients as a gastroenterologist. ZKK controlled and
confirmed the data quality, evaluated and optimized the informatics database,
wrote the paper and edited it, some other essential functions containing study
design, controlling the project and protocol development and also data
analysis. All authors revised the article carefully, read
and acknowledged the final version
of the paper.
Acknowledgment
We thank all people who were involved in this project and
contributed to us.
Conflict of interests
Authors declare
no conflict of interest.
References
1. Favoriti
P, Carbone G, Greco M, Pirozzi F, Pirozzi REM, Corcione F. Worldwide burden of
colorectal cancer: a review. Updates in surgery. 2016;68(1):7-11.
2. Kheirelseid EA, Miller N,
Chang KH, Nugent M, Kerin MJ. Clinical applications of gene expression in
colorectal cancer. Journal of gastrointestinal oncology. 2013;4(2):144.
3. Ding X, Duan H, Luo H.
Identification of core gene expression signature and key pathways in colorectal
cancer. Frontiers in genetics. 2020;11:45.
4. Graule J, Uth K, Fischer E,
Centeno I, Galván JA, Eichmann M, et al. CDX2 in colorectal cancer is an
independent prognostic factor and regulated by promoter methylation and histone
deacetylation in tumors of the serrated pathway. Clinical epigenetics.
2018;10(1):1-12.
5. Bian J, Dannappel M, Wan C,
Firestein R. Transcriptional regulation of Wnt/β-catenin pathway in colorectal
cancer. Cells. 2020;9(9):2125.
6. Mukherjee N, Dasgupta H,
Bhattacharya R, Pal D, Roy R, Islam S, et al. Frequent inactivation of MCC/CTNNBIP1
and overexpression of phospho-beta-cateninY654 are associated with breast
carcinoma: Clinical and prognostic significance. Biochimica et Biophysica Acta
(BBA)-Molecular Basis of Disease. 2016;1862(9):1472-84.
7. Katoh M. Function and cancer
genomics of FAT family genes. International journal of oncology.
2012;41(6):1913-8.
8. Wang Y, Li Z, Li W, Liu S, Han
B. Methylation of promoter region of CDX2 gene in colorectal cancer. Oncology
letters. 2016;12(5):3229-33.
9. Chang J-M, Tsai AC-D, Huang
W-R, Tseng R-C. The Alteration of CTNNBIP1 in Lung Cancer. International
journal of molecular sciences. 2019;20(22):5684.
10. Wei R, Xiao Y, Song Y, Yuan H,
Luo J, Xu W. FAT4 regulates the EMT and autophagy in colorectal cancer
cells in part via the PI3K-AKT signaling axis. Journal of Experimental &
Clinical Cancer Research. 2019;38(1):1-14.
11. Cai J, Feng D, Hu L, Chen H,
Yang G, Cai Q, et al. FAT4 functions as a tumour suppressor in gastric
cancer by modulating Wnt/β-catenin signalling. British journal of cancer.
2015;113(12):1720-9.










