Prevalence of CTX-M, OXA
and KPC genes in Klebsiella pneumoniae
isolates obtained from patients
Majid Alipour 1*, Ghoncheh
Kashani1
1
Department
of Cell and Molecular Biology, Babol Branch, Islamic Azad University, Babol,
Iran
*Corresponding Author: Majid Alipour
* Email: alipourmk@gmail.com
Abstract
Introduction: Klebsiella pneumoniae is known as one of the most important
factors in the development of opportunistic infections. The main problem in the
treatment of infections caused by these organisms is the emergence of strains
with multiple resistance, which often leads to prolonged hospital stays, increased
mortality and mobility, increased treatment costs compared to
antibiotic-sensitive microbes, and ultimately treatment failure. Therefore, the
aim of this study was to investigate the prevalence of CTX-M, OXA and KPC genes in Klebsiella
pneumoniae isolates obtained from patients.
Materials and Methods: In this study, 63 isolates of Klebsiella pneumoniae were
obtained from different clinical specimens. After final diagnosis of the
strains using standard biochemical and microbiological methods, cellular DNA
was obtained using Cinaclon's DNA extraction kit. Finally, multiplex-PCR test
was performed to evaluate the presence of OXA -48, CTX-M and KPC
genes in Eppendorf device using a pair of specific primers.
Results: Out of 63 samples under study, 29 samples (46%) from urine, 15 samples
(23.8%) from sputum, 9 samples (41.3%) from fecal samples, 5 samples (7.9%)
from wound culture and 4 samples (6.3%) were obtained from intravascular
catheter of blood culture and 1 (1.6%) sample was obtained from cerebrospinal
fluid. The results of PCR test for the studied genes showed that 49 (77.8%), 49
(77.8%) and 46 (73%) strains carried OXA, KPC and CTX-M genes,
respectively.
Conclusion: The results of this study indicate that the frequency of resistance
genes in Klebsiella pneumoniae strain is high and these strains can
transfer resistance genes with high potential to other strains. Therefore,
detection of Klebsiella pneumoniae strains containing beta-lactamase
resistance enzymes is important for better treatment and prevention of the
spread of these genes to other bacteria using accurate phenotypic and genotypic
methods.
Keywords: Klebsiella pneumoniae, Antibiotic resistance, PCR
Introduction
Klebsiella organisms are immobile and usually encapsulated. They
ferment some sugars such as lactose and sucrose. Most strains produce gas from
sugars, and the production of gas from starch is an important diagnostic
feature. Almost all grow in citrate and Moller KCN environments. These bacteria
are found in the intestines, stomachs and respiratory tract of humans and
animals. Their G + C content is 52-58% and Klebsiella pneumoniae is an indicator of this group of bacteria.
This bacterium belongs to the Enterobacteriaceae family (1). Depending on the type of
polysaccharide in the capsule, there are three capsule serological types A, B,
C in Klebsiella pneumoniae . The other three types D, E, F among Klebsiella
pneumoniae under the species Ozone
were also added to this collection and in 1949 it reached more than 8 types and
in the following years, it was renamed to letters 1, 2, 3, etc. In the
structure of these bacteria, there is O antigen, which has several different
types (2). Klebsiella pneumoniae is an important cause of community-acquired
and hospital-acquired infections. This bacterium is one of the most common
nosocomial pathogens that cause high mortality and causes various types of
infections, especially in infants, including pneumonia, sepsis, diarrhea, liver
abscess, endophthalmitis, meningitis, urinary tract infections and bacteremia.
Turns. Four million babies die every year. The highest mortality rates in
infants are related to pneumonia, sepsis, meningitis and diarrhea. Infants are
more vulnerable due to a lack of a complete immune system. Treatment of
infections in infants infected with multidrug-resistant organisms has become a
serious problem (3). Klebsiella pneumoniae can cause infections of the respiratory tract,
nasal mucosa, pharynx and pneumonia. Pneumonia is one of the major nosocomial
infections (33 to 8%). Klebsiella pneumoniae is the second most common bacterial infection
after Escherichia coli (4). It is commonly found in people
with weakened immune systems, such as hospitalized patients, diabetics, and
those with chronic lung disease. Alcoholics are often exposed to the bacterium.
Therefore, this infection comes from both the community and the hospital.
Studies in different communities show that most of the etiological causes of
urinary tract infections are bacteria of the Enterobacteriaceae family, among
which Klebsiella is the cause of 16-17% of urinary tract infections. Urinary
tract infection is one of the most common infections in all age groups that
failure to diagnose and treat in time can cause serious complications such as
urinary disorders, hypertension, renal disorders, uremia and in pregnant women
preterm delivery and even abortion (5). It is more common in women than
men. Bacteremia is one of the emergencies. In certain infections, bacteremia is
the best practical way to identify the disease-causing microorganism. Klebsiella
pneumoniae usually form
polysaccharide capsules (CPSs), also called K antigens, that cover the entire
surface of the cell. One of the main characteristics of Klebsiella
pneumoniae is the formation of
mucoid colonies on a solid culture medium. When there is a lot of carbohydrates
in the environment, the amount of polysaccharides also increases (6). Klebsiella pneumoniae develops an enzyme resistance called Klebsiella
pneumoniae carbapenemase or KPC.
Organisms containing carbapenemase can inactivate penicillin, cephalosporins,
aztreonam, and carbapenem, and because carbapenems are specifically used to
treat multidrug-resistant pathogens, resistance to this class of antibiotics is
a global threat (7). Class A carbapenemases are
phylogenetically divided into six groups, four of which are formed by the
enzymes GES, KPC, IMI / NMC, and SME, while SHV-38 and SFC each form a
separate group. Genes encoding class A carbapenemases can be located on either
the chromosome or the plasmid. Among many members of the Enterobacteriaceae
family, the genus Klebsiella containing KPC is responsible for high
rates of antibiotic resistance in recent years due to high rates of antibiotic
resistance to common antibiotics and plasmid transfer of these genes to other
species and genera of Enterobacteriaceae. Separation and identification of
strains with KPC are a real challenge for diagnostic laboratories today.
Because carbapenems are the ultimate treatment for nosocomial infections caused
by Gram-negative bacteria resistant to broad-spectrum cephalosporins, and so
far no suitable alternative to their syllables has been found. The relationship
between different studies in different parts of the world has been done in the
study of Klebsiella strains producing KPC carbapenemase. OXAcillinases
(OXAs) are also enzymes that encode resistance to carbapenems,
cephalosporins such as ceftazidime, and monobactams (aztreonam). Class D OXAcillinases
have not been extensively studied in Class A and C, but this is changing due to
the recent increase in the number of extensive clinical reports of pathogens
exhibiting OXA-related resistance. OXAcillin beta-lactamases
belong to class D in the Ambler classification, which is based on amino acid
sequences, and have serine in their active position. These beta-lactamases are
resistant to aminopenicillin and uridopenicillin (8).
The formation of mucoid colonies in Klebsiella pneumoniae is due to the presence of a thick layer of
polysaccharide capsules that can absorb large amounts of water. The
polysaccharide capsule is composed of 4 to 6 sugars, which in many cases are
uronic acid. The capsule constituents cover the bacterial surface by a thick
fibril-like structure with many layers (9). The capsule, on the one hand,
prevents the bacterium from being phagocytosed by polymorphonuclear
granulocytes, and on the other hand, from killing the bacterium by the lethal
factors of the serum. Its molecular mechanism may involve inhibiting the
activity or uptake of complement system components, particularly C3b. Recent
research has identified about 82 capsule serotypes that are antigenically
similar but different in polysaccharide skeletons. Despite antigenic diversity,
monosaccharide units are limited in number, including L-focus, L-Ramanose,
D-mannose, D-glucose, D-galactose, D-glucuronic acid, or D-galacturonic acid,
in several companions. With O-steel and pyruvate (10). Serotypes K1 and K2 in Klebsiella
pneumoniae cause liver abscess and
increase pathogenicity. It has recently been shown that K1 is a major cause of
primary liver abscess and causes metastasis, and K2 causes a secondary abscess.
Serotypes K2, K4, K5 cause community-acquired pneumonia. Capsule serotypes K2,
K7 and K33 are abundant in Klebsiella pneumoniae (11). The lipopolysaccharide (LPS)
molecule is made up of lipid A, a polysaccharide nucleus, and a side chain
called the O antigen. So far, 9 types of O antigen have been identified in Klebsiella
pneumoniae , O: 1, O: 2, O: 3, O: 4, O: 5, O: 7, O: 8, O: 9, O: 12, which
Type O: 1 is more common than other types. Natural human serum can kill
bacteria due to the complement system. The alternative pathway is activated by
bacteria in the absence of specific antibodies and plays an important role in
killing bacteria compared to the classical pathway. Lipid A also activates the
classical pathway in the absence of antibodies. Both complement pathways cause
cell lysis by the membrane attack complex and by perforating the cell wall
peptidoglycan and entering the bacterial inner membrane. Serum-resistant
bacteria contain one or a combination of polysaccharide capsules, polysaccharide
O side chains, and surface proteins (12). The wall components of
gram-negative bacteria from inside the cell to the outside include the three
main parts of the lipoprotein layer, the outer membrane, and lipopolysaccharide
(LPS).
The lipoprotein layer (Brown lipoprotein), which binds the outer
membrane layer to peptides and glycans via covalent bonding. Its protein
component consists of 57 amino acids that bind to the DAP molecule by the amino
acid lysine in the peptide tetrapeptide chain and its glycan component. Which consists of cysteine-binding thioglycerol,
binds non-covalently to the outer membrane. This lipoprotein is found in all
gram-negative bacteria except Pseudomonas aeruginosa. The outer membrane
consists of two phospholipid layers. The aim of this
study was to investigate the prevalence of CTX-M, OXA and KPC genes in Klebsiella
pneumoniae isolates obtained from patients.
Materials and Methods
In this study, 63 samples of Klebsiella pneumoniae strain were collected from patients. Due to
the importance of sterile culture media, solutions and glassware were
sterilized by autoclave at 121 ° C and 15 psi for 15 minutes. Culture media
used include Nutrient Agar, Simon Citrate, MR-VP, TSI, SIM, McConkey, EMB. Isolates
were stored in a liquid medium with glycerol at minus 20 ° C. DNA extraction using
Sinaclone DNA extraction kit (catalog number TGK1003).
The PCR was performed to identify genes using the following
conditions: initial denaturation at 94°C for 5 min, followed by 35 cycles of
denaturation (94°C for 60 s), annealing (54°C for 50 s), extension (72°C for 45
s) and final extension (72°C for 10 min) for CTX-M and KPC genes
and initial denaturation at 95°C for 5 min, followed by 35 cycles of
denaturation (95°C for 40 s), annealing (58°C for 40 s), extension (72°C for
1:30 min) and final extension (72°C for 5 min). Finally, the results were
analyzed using 1% agarose gel for electrophoresis of PCR products. All primers
used in this study are listed in Table 1.
Table 1. Sequences of primers used for evaluation of gene expression.
|
|
Sequence (5'->3') |
|
|
|
Forward primer |
Reverse primer |
|
KPC |
CGTTCTTGTCTCTCATGGCC |
CCTCGCTGTGCTTGTCATCC |
|
CTX-M |
GCGTGATACCACTTCACCTC |
TGAAGTAAGTGACCAGAATC |
|
OXA |
TTGGTGGCATCGATTATCGG |
GAGCACTTCTTTTGTGATGGC |
Statistical analysis
Graphs obtained from the data of the above experiments were
performed using EXCEL 2010 software and statistical analysis, if necessary, was
performed using SPSS 17 software. The mean of different groups was compared by
Chi-square test at the significance level of 0.05. All experiments were
performed in three replications.
Results
A total of 63 patients including 36 women (57.1%) and 27 men
(42.9%) with an age range of 16 to 71 years and a mean age of 19 4 4.1 were
included in the study.
Frequency of study samples
Out of 63 samples, 29 samples (46%) from urine, 15 samples (23.8%)
from sputum, 9 samples (41.3%) from fecal samples, 5 samples (7.9%) from wound
culture and 4 samples (6.3%) were obtained from intravascular catheter (CVP) of
blood culture and 1 (1.6%) sample were obtained from cerebrospinal fluid (CSF).
The frequency of samples collected in the present study based on gender is
shown in Table 2 and Figure 1. The results showed that out of 29 urine samples
collected, 18 samples were taken from women (62.1%) and 11 samples from men
(37.9%).
Table 2. Frequency of study samples by gender.
|
Sample type |
Female |
Male |
||
|
Number |
percentages |
Number |
percentages |
|
|
Urine |
18 |
1.62 |
11 |
9.37 |
|
sputum |
9 |
60 |
6 |
40 |
|
Feces |
4 |
4.44 |
5 |
6.55 |
|
Cultivation of wounds |
2 |
40 |
3 |
60 |
|
Blood culture |
3 |
75 |
1 |
25 |
|
CSF |
0 |
0 |
1 |
100 |

Figure 1. Results of biochemical tests of Klebsiella pneumoniae .
The results of biochemical tests of the studied strains and growth
of Klebsiella pneumoniae colonies are shown in Table 3.
Table 3. Results of biochemical tests of Klebsiella pneumoniae strains.
|
Test |
Result |
Reaction |
|
Gram staining and microscopic observation |
Gram-negative bacilli |
Red bacilli |
|
Catalase test |
Positive |
Bubble production |
|
Oxidase test |
Negative |
No change in reagent color |
|
Simon Citrate test |
Positive |
Bromothymol blue reagent color changes from green to blue |
|
TSI test |
Negative |
A / A and non-production of H2S and production of CO2
gas |
|
SIM test |
Negative |
Lack of movement and production of indole and H2S |
|
MR test |
Negative |
No red color |
|
VP test |
Positive |
Red and pink color |
|
Urease |
Positive |
Culture medium changes to pink or red |
|
Lysine decarboxylase |
Positive |
Culture medium changes to purple |
PCR reaction to evaluate the presence of OXA, KPC and
CTX-M genes
The results of PCR test for the studied genes showed that 49
(77.8%), 49 (77.8%) and 46 (73%) strains carried OXA, KPC and CTX-M
genes, respectively. Also 40 strains (63.5%) were positive for the presence of
all genes under study. No genes were observed in 5 isolates (7.9%) (Table 4).
Also, 5 (7.9%), 3 (4.8%) and 2 (3.2%) isolates, respectively, contained OXA
/ KPC, OXA / ctx and KPC / ctx genes simultaneously
(Figure 2).
Table 4. Frequency of genes.
|
Strains |
Genes |
||||||
|
OXA |
KPC |
CTX-M |
OXA/KPC |
OXA/ctx |
KPC/ ctx |
Ctx/OXA/KPC |
|
|
Number |
49 |
49 |
46 |
5 |
3 |
2 |
40 |
|
Percent |
8.77 |
8.77 |
73 |
9.7 |
8.4 |
2.6 |
5.63 |

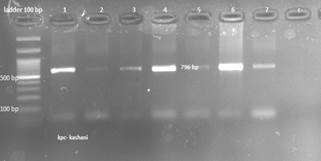

Figure 2. Electrophoresis of CTX-M, KPC and OXA gene on
1% agarose gel. C-negative control, columns 1 to 12 are the result of genes.
Discussion
Klebsiella pneumoniae or Friedlander bacillus is a
gram-negative bacillus, has a capsule, urease positive and is a member of the
Enterobacteriaceae family, which causes urinary tract infections, sepsis,
pneumonia, abdominal, pelvic, etc. The presence of resistance genes in this
bacterium has caused some therapeutic problems in recent years (13).
One of the most important antibiotics used today to treat
infections caused by Klebsiella pneumoniae is the beta-lactam group of antibiotics, but
unfortunately for reasons such as; Addition of antibiotics to the diet of
cattle, improper, excessive and arbitrary use of antibiotics and lack of strict
supervision in drug administration have led to the development of
antibiotic-resistant strains (14). The main problem in the treatment
of infections caused by these organisms is the emergence of strains with
multiple resistance, which often leads to longer hospital stays, increased
mortality and mobility, increased treatment costs compared to
antibiotic-sensitive microbes, and ultimately treatment failure (7). Beta-lactamases are enzymes that
inactivate these antibiotics by hydrolyzing the central nucleus of the
beta-lactam ring. Ambler divided these enzymes into four groups (A-D) based on
their initial structure: type B is metallobetalactamase, type C is
cephalosporinase, and type A is broad-spectrum beta-lactamase (15). 1-CTX-M belongs to the
ESBLs family and its gene is located on the plasmid and acts on cefotaxime (16). This enzyme can hydrolyze
cephalosporins and is inhibited by clavulanic acid, sulbactam and tazobactam (17). The CTX-M family of
broad-spectrum beta-lactamases was first reported from Germany in 1989 and has
since spread around the world. These enzymes are mainly present in the
Enterobacteriaceae family (18). CTX-M beta-lactamases are
not associated with TEM or HSV beta-lactamases and are only 14% similar to
these two beta-lactamases. Unlike TEM and SHV beta-lactamases, CTX-M
beta-lactamases have a greater hydrolyzing effect on cefotaxime and ceftriaxone
antibiotics than on ceftazidime (19). Carbapenems are antibiotics of the
beta-lactam family with bactericidal properties, and the cell wall of bacteria
is one of the targets of these antibiotics, which leads to disruption of
peptidoglycan synthesis (6, 10). In the present study, 63 strains
of Klebsiella pneumoniae were
collected from different clinical specimens such as blood, urine, sputum,
feces, wounds, intravascular catheter and CSF. Most of the urine samples were
isolated from women with urinary tract infections, which is consistent with the
2011 study by Salvatore et al. (20). The researchers found that urinary
tract infections were more common in women than men, meaning that more than
half of women developed UTIs at least once in their lifetime. Recurrence of the
disease is common. Risk factors associated with a high frequency of UTI
infections in women include female body anatomy, sexual intercourse, and family
history. Hashemizadeh
et al. (21) found that out of 202 collected
Klebsiella strains (180 strains of Klebsiella pneumoniae and 22 strains of Klebsiella oxytocin), 22
isolates (11.9%) carried the KPC gene. Also, in contrast to the previous
study, Aghasid Hosseini et al. (22) in Kashan in 2016 found that out of
181 collected Klebsiella, 21 (11.6%) carried the blaKPC gene, most of
which were urinary and respiratory samples from patients admitted to the ICU.
Which contradicts the present study. The results of the present study showed
that out of 63 strains under study, 49 (77.8%) strains carried the KPC
gene. Reasons for the discrepancy include geographical distance, a pattern of
drug use in the hospitals under study, and microbial genetics. On the other
hand, in agreement with the present study, Castanira et al. (23) and Chen et al. (24) found that the distribution of blaKPC
gene in Klebsiella pneumoniae strains was 76% and 73.5%, respectively. This
understanding may be due to the origin of the samples from the two studies and
the use of similar primers. In the study, Hashemizadeh et al. Found that the
origin of the samples could alter the genetic pattern of the microbe. These
researchers showed that most of the positive cases included hospital samples
(66.8%) and outpatients (33.2%). The OXA-48 gene is found in plasmids containing NDM. The
size of this plasmid is about 62.5 kb. This enzyme has been isolated from
hospitalized patients in France, Germany, Spain, and the United Kingdom, and,
as mentioned earlier, uses molecular methods to identify the gene. In the
present study, 77.8% (49 strains) carried the OXA-48 gene, which is
consistent with the study of Machuca et al. (25). In their study in Isfahan in 2020,
Solgi et al. (26) found that 66.2% of Klebsiella
pneumoniae strains carried the OXA-48
gene. Contrary to the current study, Hosseinzadeh et al. (27) found in 2018 that out of 211
strains of Klebsiella pneumoniae , only 2 isolates (0.9%) carried the OXA-48
gene. This discrepancy may be due to differences in sample type and
geographical distance. On the other hand, the results of the molecular analysis
showed that 73% (46 isolates) carried CTX-M genes, which is contrary to
the study of Lashgari et al. (28) per year. Lashgari et al. In 2014,
in a study entitled Molecular detection of beta-lactamase gene blaCTX-M
in Klebsiella pneumoniae strains
isolated from clinical samples, found that out of 100 samples of Klebsiella
pneumoniae , 46 isolates carried the gene. This discrepancy may be due to
differences in the year of the study.
Conclusion
Due to the high prevalence of Klebsiella pneumoniae and the reported resistance genes of this
bacterium, they may be mistakenly considered sensitive in routine laboratory
phenotypic tests and may develop and develop more resistant pathogens by
prescribing ineffective antibiotics. On the other hand, there is insufficient
information about the frequency of this plasmid gene and its genetic pattern in
Iran. Therefore, detection of Klebsiella pneumoniae strains containing beta-lactamase resistance
enzymes is important for better treatment and prevention of the spread of these
genes to other bacteria using accurate phenotypic and genotypic methods.
Author contributions
MA managed the project and wrote and revised the manuscript.GhK
collected the date and did the experimental tests.
Conflict of interest
The authors declare that they have no conflicts of interest.
References
1. Nordmann P, Naas T, Poirel L. Global spread of
carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791.
2. Falagas ME, Tansarli GS, Karageorgopoulos DE,
Vardakas KZ. Deaths attributable to carbapenem-resistant Enterobacteriaceae
infections. Emerg Infect Dis. 2014;20(7):1170.
3. Fraenkel-Wandel Y, Raveh-Brawer D, Wiener-Well
Y, Yinnon AM, Assous MV. Mortality due to bla KPC Klebsiella pneumoniae
bacteraemia. J Antimicrob Chemother. 2016;71(4):1083-7.
4. Wiener-Well Y, Raveh-Brawer D, Fraenkel-Wandel
Y, Yinnon AM, Assous MV. Mortality due to bla KPC Klebsiella pneumoniae
bacteraemia—authors' response. J Antimicrob Chemother. 2016;71(6):1744-.
5. Giacobbe DR, Tumbarello M, Del Bono V, Viscoli
C. Comment on: Mortality due to bla KPC Klebsiella pneumoniae bacteraemia. J
Antimicrob Chemother. 2016;71(6):1743-4.
6. Moher D, Shamseer L, Clarke M, Ghersi D,
Liberati A, Petticrew M, et al. Preferred reporting items for systematic review
and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev. 2015;4(1):1-9.
7. Daikos GL, Petrikkos P, Psichogiou M, Kosmidis
C, Vryonis E, Skoutelis A, et al. Prospective observational study of the impact
of VIM-1 metallo-β-lactamase on the outcome of patients with Klebsiella
pneumoniae bloodstream infections. Antimicrob Agents Chemother.
2009;53(5):1868-73.
8. Lodise TP, Ye M, Keyloun KR, Zhao Q, Gillard P.
Identification of patients at greatest risk for carbapenem resistance in
patients with serious hospital-onset infections due to Enterobacteriaceae
species. Open Forum Infect Dis. 2016;3(suppl_1):1794.
9. Walther-Rasmussen J, Høiby N. Class A
carbapenemases. J Antimicrob Chemother. 2007;60(3):470-82.
10. Vardakas KZ, Rafailidis PI, Konstantelias AA,
Falagas ME. Predictors of mortality in patients with infections due to multi-drug
resistant Gram negative bacteria: the study, the patient, the bug or the drug?
J Infect. 2013;66(5):401-14.
11. Castón JJ, Lacort-Peralta I, Martín-Dávila P,
Loeches B, Tabares S, Temkin L, et al. Clinical efficacy of
ceftazidime/avibactam versus other active agents for the treatment of
bacteremia due to carbapenemase-producing Enterobacteriaceae in hematologic
patients. Int J Infect Dis. 2017;59:118-23.
12. Ben-David
D, Kordevani R, Keller N, Tal I, Marzel A, Gal-Mor O, et al. Outcome of carbapenem
resistant Klebsiella pneumoniae bloodstream infections. Clin Microbiol Infect.
2012;18(1):54-60.
13. Karaiskos I, Giamarellou H.
Multidrug-resistant and extensively drug-resistant Gram-negative pathogens:
current and emerging therapeutic approaches. Expert Opin Pharmacother.
2014;15(10):1351-70.
14. Pitout JD, Nordmann P, Poirel L.
Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global
nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873-84.
15. Liu S-W, Chang H-J, Chia J-H, Kuo A-J, Wu T-L,
Lee M-H. Outcomes and characteristics of ertapenem-nonsusceptible Klebsiella
pneumoniae bacteremia at a university hospital in Northern Taiwan: a matched
case-control study. J Microbiol Immunol Infect. 2012;45(2):113-9.
16. Bartoletti M, Giannella M, Caraceni P,
Domenicali M, Ambretti S, Tedeschi S, et al. Epidemiology and outcomes of
bloodstream infection in patients with cirrhosis. J Hepatol. 2014;61(1):51-8.
17. Tumbarello M, Viale P, Viscoli C, Trecarichi
EM, Tumietto F, Marchese A, et al. Predictors of mortality in bloodstream
infections caused by Klebsiella pneumoniae carbapenemase–producing K.
pneumoniae: importance of combination therapy. Clin Infect Dis.
2012;55(7):943-50.
18. DerSimonian R, Laird N. Meta-analysis in
clinical trials. Control Clin Trials. 1986;7(3):177-88.
19. Gallagher JC, Kuriakose S, Haynes K, Axelrod
P. Case-case-control study of patients with carbapenem-resistant and
third-generation-cephalosporin-resistant Klebsiella pneumoniae bloodstream
infections. Antimicrob Agents Chemother. 2014;58(10):5732-5.
20. Salvatore S, Salvatore S, Cattoni E, Siesto G,
Serati M, Sorice P, et al. Urinary tract infections in women. Eur J Obstet
Gynecol Reprod Biol. 2011;156(2):131-6.
21. Mirnejhad R, Hashemizadeh FS, Zamanzad B,
Jahandideh S, Ansari N, Gholipour A, et al. Identification of KPC-producing
Klebsiella pneumoniae in clinical samples in Iran. Yafte journal of medical
scienes (YJMS). 2013;15(1).
22. Agha-Seyed Hosseini M, Firoozeh F, Piroozmand
A, Gilasi HR. Carbapenemase-producing Klebsiella pneumoniae strains among
clinical specimens in Kashan (2014-2015). Feyz. 2016;20(3):267-73.
23. Castanheira M, Sader HS, Deshpande LM,
Fritsche TR, Jones RN. Antimicrobial activities of tigecycline and other
broad-spectrum antimicrobials tested against serine carbapenemase-and
metallo-β-lactamase-producing Enterobacteriaceae: report from the SENTRY
Antimicrobial Surveillance Program. Antimicrob Agents Chemother.
2008;52(2):570-3.
24. Chen S, Hu F, Xu X, Liu Y, Wu W, Zhu D, et al.
High prevalence of KPC-2-type carbapenemase coupled with CTX-M-type
extended-spectrum β-lactamases in carbapenem-resistant Klebsiella pneumoniae in
a teaching hospital in China. Antimicrob Agents Chemother. 2011;55(5):2493-4.
25. Machuca J, López-Cerero L, Fernández-Cuenca F,
Mora-Navas L, Mediavilla-Gradolph C, López-Rodríguez I, et al.
OXA-48-like-producing Klebsiella pneumoniae in Southern Spain in 2014–2015.
Antimicrob Agents Chemother. 2019;63(1):e01396-18.
26. Solgi H, Nematzadeh S, Giske CG, Badmasti F,
Westerlund F, Lin Y-L, et al. Molecular epidemiology of OXA-48 and NDM-1
producing enterobacterales species at a University Hospital in Tehran, Iran,
between 2015 and 2016. Front Microbiol 2020;11:936.
27. Hosseinzadeh Z, Ebrahim-Saraie HS, Sarvari J,
Mardaneh J, Dehghani B, Rokni-Hosseini SMH, et al. Emerge of bla NDM-1 and bla
OXA-48-like harboring carbapenem-resistant Klebsiella pneumoniae isolates from
hospitalized patients in southwestern Iran. J Chin Med Assoc. 2018;81(6):536-40.
28. Lashgari N, Vand Yousefi J, Siadat SD,
Shahcheraghi F, Khosravi M, Vakili H, et al. Identification of bla-CTX-M
β-lactamase in Klebsiella pnumoniae clinical isolates by polymerase chain
reaction. Med Sci J Islamic Azad Univ Tehran Med Branc. 2014;24(3):148-52.