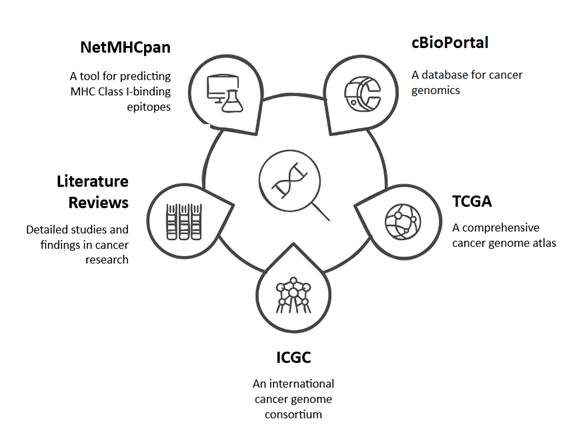
Figure 1. Schematic of selected potential tumor antigens of renal cell carcinoma.
Exploring neoantigens and genetic profiles in renal
cell carcinoma: a study of Iranian patients
Zohreh Mehmandoostli
1,2, Mahmood Dehghani Ashkezari 1,2,
Seyed Morteza Seifati 1,2, Amirreza Farzin 3, Gholam Ali Kardar 4,5*
1 Department of Biology, Ashk.C., Islamic
Azad University, Ashkezar, Iran
2 Medical Biotechnology Research Center, Ashk.C.,
Islamic Azad University, Ashkezar, Iran
3 Uro-Oncology Research Center, Tehran University of Medical
Sciences, Tehran, Iran
4 Immunology, Asthma and Allergy Research Institute (IAARI), Tehran University
of Medical Sciences, Tehran, Iran
5 Department of Medical Biotechnology, School of Advanced
Technologies in Medicine, Tehran University of Medical Sciences, Tehran, Iran
* Corresponding Author:
Gholam Ali Kardar
* Email: gakardar@tums.ac.ir
Abstract
Introduction: Kidney cancer
accounts for 3.7% of all newly diagnosed cancer cases and represents a
considerable global health challenge. Although there have been advancements in
treatment, renal cell carcinoma (RCC) continues to show resistance to
conventional cytotoxic chemotherapy, highlighting the need for innovative
therapeutic strategies. Current research is focusing on vaccine approaches that
target tumor neoantigens, utilizing next-generation sequencing to pinpoint
tumor-specific mutations. A deeper understanding of the molecular
characteristics of RCC, particularly gene mutations such as BAP1, PBRM1, SETD2,
and VHL, is essential for the development of personalized treatment modalities.
This study aimed to investigate potential tumor neoantigens in samples from Iranian
patients diagnosed with RCC, with an emphasis on peptide sequences that exhibit
a strong binding affinity for Iranian Human Leukocyte Antigen (HLA).
Materials and
methods: Databases and
relevant literature were employed to identify neoantigens with the highest
prevalence. Tumor samples were obtained from patients with RCC, and primary
cells were isolated and cultured in RPMI complete medium. Total DNA was
extracted, followed by polymerase chain reaction (PCR) using specifically
designed primers, and the resulting PCR products were sequenced using Sanger
sequencing.
Results: Our examination did not identify the specified nucleotide changes in
the DNA sequencing chromatogram of the cell samples, indicating that the
anticipated mutations were absent. Nevertheless, other mutations were observed
in the analyzed regions of the genes.
Conclusion: Although certain mutations were not identified in the sequenced
samples, this research highlights the necessity for further investigation.
Comprehensive studies are vital to gain a complete understanding of the genetic
mutation profile of RCC in Iranian patients. Mapping the gene mutation
landscape among RCC patients in Iran presents significant opportunities for the
development of effective cancer vaccines and tailored treatment strategies.
Keywords: Human Leukocyte Antigen (HLA), Renal cell carcinoma, Tumor, Mutation,
Vaccines
Introduction
Kidney cancer represents 3.7% of newly diagnosed cancer cases and
ranks among the ten most prevalent cancers. The predominant type, RCC, is found
in 85% of kidney cancer instances. This condition is more prevalent in men than
in women, with a ratio of 1.7:1, and it generally affects older adults, with an
average age of 64 at diagnosis (1). The rate of metastasis is notably high, as 30% of patients are
diagnosed with metastatic disease, and an additional 30% develop metastases
during subsequent follow-ups (2). Renal cell carcinomas can be categorized into clear cell (CCRCC),
papillary (PRCC), and chromophobe (ChRCC) subtypes,
which account for 65-70%, 15-20%, and 5-7% of all RCC cases, respectively (3). Recent research has identified genetic factors, including
mutations in the VHL, PBRM1, BAP1, and SETD2 genes, as significant contributors
to the risk of developing RCC (4).
Among the
innovative strategies for treating RCC, antigen-directed approaches such as
monoclonal antibodies, adoptive cell therapies, and therapeutic vaccines show
promise in enhancing the effectiveness of existing immunotherapies (5). Various neoantigen vaccine platforms, including synthetic long
peptide (SLP), RNA, dendritic cell (DC), and DNA vaccines, are currently
undergoing evaluation in early-phase clinical trials (6). The SLP vaccine platform has emerged as the most extensively
studied neoantigen vaccine in preclinical and early-phase clinical settings,
offering notable advantages such as a well-established safety profile, a
thoroughly characterized GMP manufacturing process, excellent stability, and
ease of administration in human trials (7). The advent of next-generation sequencing (NGS) technologies has
further expedited the discovery of tumor-specific mutations, facilitating the
development of more targeted and personalized treatment strategies (8).
Recent studies
have validated the therapeutic promise of SLP neoantigen vaccines across
various human cancers, including melanoma, glioblastoma, non-small cell lung
cancer (NSCLC), colorectal cancer, and urothelial cancer (9-13). A clinical trial involving the peptide-based neoantigen vaccine
iNeo-Vac-P01 in patients with advanced solid tumors revealed a favorable safety
profile and encouraging efficacy, achieving a 71.4% disease control rate and
robust immune responses in nearly 80% of participants (9). In this context, Ott et al. have undertaken multiple
investigations. In one study involving six melanoma patients, clinicians
administered up to 20 neoantigens per individual. Remarkably, four patients
with stage IIIB/C disease experienced no recurrence over a 25-month period,
while two patients with previously untreated stage IVM1b disease (lung
metastases) achieved complete regression following vaccination and subsequent
anti-PD-1 therapy (12). Similar findings were reported in glioblastoma patients (11). Furthermore, Ott et al. introduced a neoantigen-based vaccine,
NEO-PV-01, in conjunction with PD-1 blockade for patients with melanoma, NSCLC,
or bladder cancer. This vaccine elicited CD4+ and CD8+ T cell responses
post-vaccination, characterized by a cytotoxic phenotype capable of
infiltrating tumors and inducing tumor cell destruction. The treatment was
deemed safe, with no adverse events reported (NCT02897765) (13).
In tumor cells,
mutations can lead to the formation of novel self-antigen epitopes, referred to
as neoantigens. The advent of NGS has created opportunities for the efficient
and cost-effective identification of tumor-specific mutations in individual
patients, facilitating the exploration of therapies targeting these mutated
proteins in clinical trials studies (14). This process encompasses several stages, including the collection
of tumor samples, sequencing, bioinformatic analysis, and peptide synthesis,
all of which demand considerable time and resources. Despite advancements in
sequencing technology facilitating rapid progress, the identification and
validation of neoantigens remain labor-intensive and costly, with the
preparation of vaccines from tissue samples typically requiring 3 to 5 months (15, 16).
In RCC, the
scenario is different because RCC is a cancer with low mutational burden (17).
Neoantigens can be divided into two categories: shared neoantigens (public
neoantigens) and personalized neoantigens. Off-the-shelf vaccines are designed
to target shared neoantigens, which are anticipated to have a high expression
frequency and provoke strong anti-tumor immune responses, thus making them
suitable for a wider array of cancer specially RCC patients (18). The molecular profiles of RCC have been examined through NGS in
various research initiatives, including The Cancer Genome Atlas (TCGA) and
other studies conducted in France, Japan, and the European Union (19). Wang et al. analysis of
individual gene mutations was shown that the most prevalent mutations include
PBRM1 (74.1%), BAP1 (74.1%), SETD2 (74.1%), and VHL (74.91%) (20). In our forthcoming research aimed at developing an off-the-shelf
peptide vaccine, we explored potential tumor neoantigens in RCC samples
collected from Iranian patients, focusing on peptide sequences with a strong
affinity for the Iranian HLA.
Materials and methods
3.1. Data
sources
A genomic analysis of RCC data was performed using the TCGA database (https://portal.gdc.cancer.gov/). Literature reviews and original articles were also investigated to
find RCC mutations. The potential neoantigens were explored using cBioPortal (http://www.cbioportal.org, v3.2.11). The Human Gene Mutation
Database (HGMD, www.hgmd.com) was used to select peptide sequences considering a moderate to strong
affinity for Iranian HLA binding. The bioinformatics analysis included the
prediction of MHC I-binding epitopes using tools such as NetMHCpan
(v4.0) to identify peptides with a moderate to high affinity for HLA molecules.
3.2. Patients
and samples
Cancerous tissues were obtained from patients diagnosed with RCC
following surgical procedures at the Department of Urology at Imam Khomeini
Hospital. Pathologists verified the pathological features of these samples
through histopathological analysis. The section on Ethical Considerations
outlines the ethical approval for utilizing the primary cells of patients, with
written informed consent secured from all participants (Ethical Approval Code:
IR.TUMS.IAARI.REC.1399.010). The inclusion criteria specified that participants
must be diagnosed with RCC and have not received any prior treatment. Patients
with different histological subtypes of RCC or those who had previously
undergone treatment were excluded from the study.
3.3. Primary Cell
Culture
RCC solid tumor tissue was first rinsed
with phosphate-buffered saline (PBS) and then cut into pieces measuring
approximately 1-2 mm. The tissue was subsequently filtered through a 70 μm cell strainer without the use of enzymatic
digestion. The resulting filtered cells were washed twice with PBS and cultured
in RPMI 1640 medium supplemented with 15% fetal bovine serum (FBS) and 1X
Penicillin-Streptomycin (Pen/Strep) in a 37°C incubator with 5% CO2 and 95%
humidity for a duration of 72 hours. Following this incubation, the suspended
cells were removed by washing with sterile PBS. The adherent cells were then
maintained in fresh RPMI medium containing 10% FBS and 1X Pen/Strep for one
week, allowing them to achieve 90% confluency. The cells were subsequently passaged
using Trypsin-EDTA (0.25%) and seeded at a density of 1x106 cells
per T-75 flask for further experimentation.
3.4. DNA Extraction and Polymerase Chain Reaction
To validate the potential neoantigen peptides in cultured cells,
adherent cells were harvested. Total DNA was extracted utilizing the SanPrep Column DNA Miniprep Kit (Bio Basic,
Canada) in accordance with the manufacturer's guidelines. The PCR was performed
using Taq DNA Polymerase 2x Master Mix and 1.5 mM MgCl2 (Ampliqon,
Denmark), following the standard protocol with an annealing temperature of 58°C
for the primers. Specific primers for the peptides were designed using Primer3.
To ensure the specificity and accuracy of the designed primers, their sequences
were subjected to a BLAST search in NCBI and Gene Runner. The sequences of the
primers are provided in Table 1.
Table 1. The details of oligonucleotide primers
utilized for PCR.
|
Reverse Primer |
Forward Primer |
Gene |
|
|
525 bp |
GCTTCAGACCGTGCTATCGT |
GCGTTCCATCCTCTACCGAG |
VHL (1) |
|
334 bp |
ACGTACAAATACATCACTTCCATTT |
ACCGGTGTGGCTCTTTAACA |
VHL (2) |
|
579 bp |
CGATATGCTGCAATTCCCACT |
GGCAAAGCCTCTTGTTCGTT |
VHL (3) |
|
217 bp |
CCATGCTGGAGTACAGTGAGTT |
TGATGCACATATCCTGGAGAAGTTA |
PBRM1 |
|
555 bp |
AGCAGTTGAGCCAGGGAATC |
GCTGCTCTCTGAAGCTTTGC |
BAP1 |
|
310 bp |
TTAATGGTCAGAACAGCAATCGTG |
AAACTTTGAAGCTGGTAGTCAGGA |
SETD2 |
|
167 bp |
GAGGCTGGGGCACAGCAGGCCAGTG |
CTCCTAGGTTGGCTCTG |
TP53 |
|
451 bp |
CCCCTGAGAAGCAGTCTGTG |
GCTGGGGAGGTTTCATGGAG |
FLCN |
|
250bp |
CTGAGATCAGCCAAATTCAGTT |
GGGAAAAATATGACAAAGAAAGC |
PIK3CA (1) |
|
241bp |
TGGAATCCAGAGTGAGCTTTC |
CTCAATGATGCTTGGCTCTG |
PIK3CA (2) |
3.5. Analysis
of PCR Product
The PCR products underwent sequencing at Pishgam
Co. in Iran, utilizing the Sanger sequencing technique. Following the
amplification of the selected mutant regions with the specified primer sets
(refer to Table 1), a total of 10 PCR product samples were dispatched for
sequencing. The sequencing chromatograms were first analyzed using Chromas Lite
v.2.5 software, followed by alignement and comparison
to reference sequences using CLC Sequence Viewer 7.7.1 software to investigate
the potential presence of neoantigen mutations in the cultured primary tumor
cells.
Ethical
consideration
The Vice Chancellor for Research Affairs at Tehran University of
Medical Sciences in Tehran, Iran, granted approval for the acquisition of the
primary cell culture utilized in this study (Ethical Approval Code:
IR.TUMS.IAARI.REC.1399.010). All procedures conducted adhered to the ethical
standards set forth by both the institutional and national research committees,
as well as the 1964 Helsinki Declaration and its subsequent amendments or
equivalent ethical guidelines.
Data Analysis
Statistical methods were not applied in the analysis of sequencing
data. Instead, descriptive statistics were utilized to summarize the frequency
and distribution of the identified neoantigens. Additionally, bioinformatic
tools, including NetMHCpan 4.1, were employed to
predict the binding affinity of the identified peptides to HLA molecules.
Results
4.1.
Bioinformatic Data
The potential tumor antigens associated with RCC were identified
through the utilization of cBioPortal, The Cancer
Genome Atlas (TCGA), the International Cancer Genome Consortium (ICGC) cohorts,
and comprehensive literature reviews. Following the identification of the most
prevalent antigens exhibiting mutations, involve of 17 mutations from 7 genes
(VHL, PBRM1, BAP1, SETD2, TP53, FLCN, and PIK3CA). By advantage of
bioinformatics tools, including NetMHCpan (v4.1),
MHC-I binding epitopes predication, common Iranian population MHC-I are
HLA-B*35:01, HLA-A*02:01, HLA-A*24:02, HLA-B*51:01, HLA-A*03:01, and
HLA-A*01:01, in both wild-type and mutant of 1105 nine-mer
peptides were done. Mutant peptides with moderate to high binding affinities in
compare to wild type were selected based on their predicted binding
capabilities. Finally, based on Table 2, twenty nine peptides were selected (15,21).
A summary of the RCC potential tumor antigens data, along with the
common MHC-I alleles in the Iranian population, is presented in Figure 1 and
Table 2.


Figure 1. Schematic of selected potential tumor antigens of renal cell carcinoma.
Table 2. The details of selected potential tumor
antigens of Renal Cell Carcinoma. The peptides with strong binding affinity
to HLA were shown with green color. peptides exhibiting moderate to strong
HLA-binding affinity (IC50 < 150 nmol/l) being more likely to activate CD8+
T cells.
|
Gene |
Mutation |
Immunizing peptide |
HLA |
Binding Affinity (nM) |
|
|
Wild |
Mutant |
||||
|
VHL |
W88L |
SPRVVLPVL |
HLA-B*35:01 |
3839.92 |
1613.19 |
|
VVLPVLLNF |
HLA-A*24:02 |
376.32 |
1420.44 |
||
|
H115Y |
SYRGYLWLF |
HLA-A*24:02 |
14.56 |
12.14 |
|
|
D121Y |
RYAGTHDGL |
HLA-A*24:02 |
26237.97 |
387.69 |
|
|
V130F |
GLLFNQTEL |
HLA-A*02:01 |
176.25 |
108.44 |
|
|
LLFNQTELF |
HLA-A*24:02 |
4051.01 |
2081.94 |
||
|
LLFNQTELF |
HLA-B*35:01 |
1934.56 |
1456.79 |
||
|
N131K |
GLLVKQTEL |
HLA-A*02:01 |
176.25 |
197.59 |
|
|
GTHDGLLVK |
HLA-A*03:01 |
22426.29 |
342.46 |
||
|
LLVKQTELF |
HLA-A*24:02 |
4051.01 |
2578.81 |
||
|
LLVKQTELF |
HLA-B*35:01 |
1934.56 |
2834.24 |
||
|
L135F |
LVNQTEFFV |
HLA-A*02:01 |
736.1 |
140.49 |
|
|
LLVNQTEFF |
HLA-B*35:01 |
1934.56 |
1702.89 |
||
|
I151T |
FANTTLPVY |
HLA-A*01:01 |
2066.66 |
1207.22 |
|
|
TTLPVYTLK |
HLA-A*03:01 |
31.53 |
39.98 |
||
|
FANTTLPVY |
HLA-B*35:01 |
4.85 |
3.96 |
||
|
L169P |
LQVVRSPVK |
HLA-A*03:01 |
807.91 |
1784.2 |
|
|
PBRM1 |
G626V |
VPLPDDDDM |
HLA-B*35:01 |
416.12 |
58.65 |
|
BAP1 |
F170V |
RTMEAFHVV |
HLA-A*02:01 |
7.22 |
29.91 |
|
RTMEAFHVV |
HLA-A*24:02 |
588.39 |
703.75 |
||
|
MEAFHVVSY |
HLA-B*35:01 |
59.51 |
62.01 |
||
|
EAFHVVSYV |
HLA-B*51:01 |
2045.28 |
1749.79 |
||
|
SETD2 |
H1629Y |
NMYSCEPNC |
HLA-A*02:01 |
12006.73 |
3790.55 |
|
TP53 |
R248L |
NLRPILTII |
HLA-A*02:01 |
26600.99 |
3128.28 |
|
MNLRPILTI |
HLA-A*24:02 |
9934.93 |
3516.92 |
||
|
FLCN |
R18H |
ELHGPHTLF |
HLA-A*24:02 |
16730.88 |
8074.98 |
|
ELHGPHTLF |
HLA-B*35:01 |
13223.15 |
9173.56 |
||
|
PIK3CA |
E542K |
ISTRDPLSK |
HLA-A*03:01 |
35089.89 |
930.82 |
|
H1047L |
ALHGGWTTK |
HLA-A*03:01 |
5388.83 |
47.14 |
|
4.2.
Pathological Characteristics of RCC
The identification of renal cell tumor was validated through
Hematoxylin and eosin (H&E) staining (Fig.1). This staining technique
allowed for a comprehensive visualization of cellular morphology and tissue
structure, thereby affirming the presence of distinctive characteristics
associated with RCC. Following a one-week culture period, RCC cells were
analyzed using an inverted microscope to assess their morphology and growth
patterns (Fig.2). The cells displayed the typical morphological traits of RCC,
characterized by clear cytoplasm and well-defined cell boundaries. Finally, 10
RCC patient cells were selected for DNA sequencing.
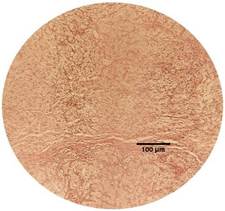
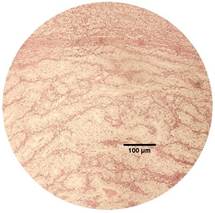
Figure 2. Two different RCC tissues from the studied patient #008 were stained
with the hematoxylin-eosin staining method.
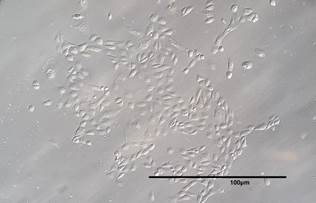
Figure 3. Primary RCC cell obtained from patient #004 (20X magnification).
Table 3. The patient demographics of those with RCC who used their tumor samples
for the study.
|
Patient No. |
Sex |
Age (Years) |
Diagnosis |
|
01 |
Female |
46 |
Clear cell |
|
02 |
Male |
53 |
Chromophobe |
|
03 |
Male |
41 |
Clear cell |
|
04 |
Female |
48 |
Clear cell |
|
05 |
Male |
39 |
Papillary |
|
06 |
Female |
46 |
Clear cell |
|
07 |
Male |
71 |
Clear cell |
|
08 |
Male |
45 |
Clear cell |
|
09 |
Male |
66 |
Papillary |
|
10 |
Female |
74 |
Chromophobe |
4.3 Sanger
Sequencing Analysis
The results from Sanger sequencing revealed that the expected mutations
were absent in the analyzed samples when compared to the reference sequences.
However, additional mutations in the analyzed genes, which are not the chosen
mutations, were observed. Below are some of those mutations found in the
sequenced regions that include: PIK3CA p.Leu1067del,
VHL L135I, VHL c.491delA (p.Gln164ff), TP53 D259E, …
This discrepancy indicates that the identified neoantigens may not be
common among Iranian RCC patients. To validate these findings, additional
sequencing with a larger sample size may be required.
Discussion
The global
incidence of renal cell carcinoma (RCC) is on the rise, with an estimated
mortality rate of approximately 20% (22). Neoantigen-based vaccines may prove effective for patients
suffering from metastatic RCC, potentially leading to improved overall survival
rates, diminished tumor burden, and a reduction in cancer progression (23). It is crucial to identify tumor-associated antigens that are
specific to RCC and to develop strategies to counteract tumor-induced
immunosuppression, thereby making vaccine therapy a feasible treatment option (24). The ideal antigen should be exclusive to tumor cells, vital for
tumor initiation and progression, identifiable by the immune system, and
capable of inducing cytotoxic effects on tumor cells (25). Although numerous tumor antigens show promise, only a limited
number meet the criteria for tumor selectivity and robust immune response
activation, underscoring the necessity for ongoing research and development in
this field (26). Previous investigations have confirmed the safety and feasibility
of employing mutated VHL peptides as a vaccine for metastatic RCC (27). In the majority of ccRCC cases,
mutations in genes such as SETD2, BAP1, MTOR, PTEN, KDM5C, and PBRM1 are
frequently observed in the chromosome 3p regions, alongside VHL gene mutations (28). Recent research has emphasized the potential of these mutations
as targets for innovative therapeutic strategies, indicating that a
multi-targeted vaccine approach may enhance the efficacy of RCC immunotherapy.
In a study conducted by Razafinjatovo et al. in 2017,
DNA samples from 30 ccRCC patients were analyzed
through NGS, focusing on a specific panel of 18 genes, including VHL, BAP1,
HIF1a, PDGFRA, PDGF(R)B, TP53, CARD11, NFkB, TSC1,
MTOR, EGFR, PBRM1, SETD2, KDM5C, KDM6A, PTEN, and PIK3CA, all of which are
known to harbor mutations in ccRCC. Frequent
mutations in these genes were detected, and their mutational status,
particularly in genes such as PBRM1, BAP1, CARD11, and HIF1α, was
correlated with responses to targeted therapies, potentially serving as
predictors of therapeutic outcomes in ccRCC patients (29).
These findings
have established a basis for the identification of potential tumor antigens.
The advent of advanced sequencing technologies has facilitated more accurate
and thorough analyses of these mutations, thereby deepening our comprehension
of their significance in RCC. Such insights are vital for the creation of
targeted vaccines and personalized therapies that can effectively address the
genetic diversity present in RCC tumors. Recent investigations have indicated
that neoantigen-based vaccines administered to RCC patients have shown
encouraging outcomes. Nevertheless, two major challenges persist in this area.
The first challenge is the impracticality of a peptide-based approach due to
prohibitive costs and time demands (30). For example, while a particular study reported positive outcomes,
the process was impeded by these financial and temporal constraints (12). The application of NGS techniques for personalized treatment and
the identification of neoantigens may prove to be highly effective. Research
conducted by Bles and Ott (2021) has illustrated that personalized
neoantigen-based vaccines, supported by rapid sequencing technologies and
bioinformatics, exhibit strong immunogenic profiles and preliminary indications
of anti-tumor efficacy in melanoma patients. Furthermore, findings from Ott et
al. (2017) highlight that these vaccines can provoke robust anti-tumor immune
responses and contribute to tumor size reduction in certain melanoma patients (31, 32).
In this
context, the exploration of universal peptides as an alternative strategy is
particularly compelling. Universal peptides are present across a wide range of
cancer types and have the unique ability to stimulate immune responses against
multiple tumors simultaneously. This capacity enables them to activate various
subsets of T cells, positioning universal peptides as promising candidates for
broad-spectrum vaccines in cancer therapy. The NeoVax
brand is focused on identifying and utilizing these universal peptides to cater
to different cancer types. By adopting this approach, we can significantly
reduce the time and costs associated with the collection of individual tumor
samples while ensuring a consistent method of application. This could provide a
hopeful pathway for addressing the challenges faced in the treatment of RCC and
other cancers.
However, there
remains a substantial gap in research specifically addressing the application
of these vaccines for renal cell carcinoma. This shortfall may be attributed to
the distinct genetic characteristics of RCC, which exhibit unique mutational
patterns different from those seen in melanoma. Moreover, the tumor
microenvironment in RCC can significantly impact the immune system’s ability to
recognize and respond to neoantigens.
In our
research, we carefully examined a variety of potential tumor antigens from
samples collected from Iranian patients, as outlined in Table 2. While we
started with optimistic expectations, we found that we could not validate the
mutations in the sequenced samples from Iran. This lack of confirmation might
be influenced by several factors, including sample heterogeneity and varying
mutation frequencies among patients in this population. Future investigations
should prioritize the use of higher coverage sequencing methods and an expanded
sample size to address these challenges. Furthermore, incorporating additional
techniques like whole exome sequencing (WES) or RNA sequencing (RNA-seq) could
yield a more thorough understanding of the genetic landscape of RCC across
diverse populations(33, 34). The genetic profile of renal tumors exhibits spatial variation
within individual tumors as well as differences among patients. There is a
notable deficiency in data concerning the intratumor heterogeneity present
across various racial groups. Preliminary evidence indicates that genetic
variations may exist among different racial demographics affected by RCC,
warranting further investigation (35). For instance, analysis of the TCGA database reveals that black
patients demonstrate elevated expression of BAP1 at the genetic level (36). Future studies should focus on clarifying the degree of genetic
heterogeneity within RCC tumors and its potential implications for personalized
treatment strategies. Gaining insight into these variations is crucial for the
development of targeted therapies that are more effective for distinct racial
and ethnic populations.
Conclusion
Author
contribution
ZM, GAK, formal analysis, investigation, data curation,
visualization, and writing original draft preparation. GAK,
conceptualization, methodology, validation, and project administration. GAK,
writing review and editing, supervision and funding acquisition. AF made
significant contributions to the preparation of the primary cells. MDA
contributed to the study and problem-solving related to the paper, writing
review and editing. SMS contributed to writing, reviewing and editing.
All authors read and approved the final manuscript.
Ethics approval
The author
declares no conflict of interest.
Funding
The ethical
committee of Tehran University of Medical Science confirmed this study [Ethic
no. IR.TUMS.IAARI.REC.1399.010].
Consent for
publication
All authors
have reviewed and approved the final manuscript for publication. All
participants provided informed consent for their data and images to be
published.
Funding
Tehran
University of Medical Science (Grant No. 48430) supported this work. The
authors declare that no funds, grants, or other support were received during
the preparation of this manuscript.
References
1. Mirlekar B,
Pylayeva-Gupta YJC. IL-12 family cytokines in cancer and immunotherapy.
Cancers. 2021;13(2):167.
2. Deleuze A, Saout J,
Dugay F, Peyronnet B, Mathieu R, Verhoest G, et al. Immunotherapy in renal cell
carcinoma: the future is now. International Journal of Molecular Sciences.
2020;21(7):2532.
3. Inamura K. Renal cell
tumors: understanding their molecular pathological epidemiology and the 2016
WHO classification. International journal of molecular sciences.
2017;18(10):2195.
4. Carril-Ajuria L, Santos
M, Roldán-Romero JM, Rodriguez-Antona C, de Velasco GJC. Prognostic and
predictive value of PBRM1 in clear cell renal cell carcinoma. Cancers (Basel).
2019;12(1):16.
5. Braun DA, Bakouny Z,
Hirsch L, Flippot R, Van Allen EM, Wu CJ, et al. Beyond conventional
immune-checkpoint inhibition—novel immunotherapies for renal cell carcinoma.
Nature reviews Clinical oncology. 2021;18(4):199-214.
6. Supabphol S, Li L,
Goedegebuure SP, Gillanders WE. Neoantigen vaccine platforms in clinical
development: understanding the future of personalized immunotherapy. Expert
opinion on investigational drugs. 2021;30(5):529-41.
7. Blum JS, Wearsch PA,
Cresswell P. Pathways of antigen processing. Annual review of immunology.
2013;31:443-73.
8. Morganti S, Tarantino
P, Ferraro E, D’Amico P, Duso BA, Curigliano GJTr, et al. Next generation
sequencing (NGS): a revolutionary technology in pharmacogenomics and
personalized medicine in cancer. Adv Exp Med Biol. 2019:9-30.
9. Fang Y, Mo F, Shou J,
Wang H, Luo K, Zhang S, et al. A pan-cancer clinical study of personalized
neoantigen vaccine monotherapy in treating patients with various types of
advanced solid tumors. Clinical Cancer Research. 2020;26(17):4511-20.
10. Hilf N, Kuttruff-Coqui
S, Frenzel K, Bukur V, Stevanović S, Gouttefangeas C, et al. Actively
personalized vaccination trial for newly diagnosed glioblastoma. Nature.
2019;565(7738):240-5.
11. Keskin DB, Anandappa AJ,
Sun J, Tirosh I, Mathewson ND, Li S, et al. Neoantigen vaccine generates
intratumoral T cell responses in phase Ib glioblastoma trial. Nature.
2019;565(7738):234-9.
12. Ott PA, Hu Z, Keskin DB,
Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine
for patients with melanoma. Nature. 2017;547(7662):217-21.
13. Ott PA, Hu-Lieskovan S,
Chmielowski B, Govindan R, Naing A, Bhardwaj N, et al. A phase Ib trial of
personalized neoantigen therapy plus anti-PD-1 in patients with advanced
melanoma, non-small cell lung cancer, or bladder cancer. Cell.
2020;183(2):347-62. e24.
14. Blass E, Ott PA.
Advances in the development of personalized neoantigen-based therapeutic cancer
vaccines. Nature Reviews Clinical Oncology. 2021;18(4):215-29.
15. Peng M, Mo Y, Wang Y, Wu
P, Zhang Y, Xiong F, et al. Neoantigen vaccine: an emerging tumor
immunotherapy. Molecular Cancer. 2019;18:1-14.
16. Chong C, Coukos G,
Bassani-Sternberg MJNb. Identification of tumor antigens with
immunopeptidomics. Nat Biotechnol. 2022;40(2):175-88.
17. Braun D.A, Moranzoni G,
Chea V, McGregor BA,
Blass E,Tu CR. A neoantigen vaccine generates antitumor
immunity in renal cell carcinoma. Naturer. 2025;639: 474-482.
18. Li X, You J, Hong L, Liu
W, Guo P, Hao X. Neoantigen cancer vaccines: a new star on the horizon. Cancer
Biology & Medicine. 2024;21(4):274.
19. Wang J, Xi Z, Xi J,
Zhang H, Li J, Xia Y, et al. Somatic mutations in renal cell carcinomas from
Chinese patients revealed by whole exome sequencing. Cancer Cell International.
2018;18(1):1-12.
20. Wang Y, He P, Zhou X,
Wang C, Fu J, Zhang D, et al. Gene mutation profiling and clinical
significances in patients with renal cell carcinoma. Clinics. 2023;78:100259.
21. Fritsch EF, Rajasagi M,
Ott PA, Brusic V, Hacohen N, Wu CJ. HLA-binding properties of tumor neoepitopes
in humans. Cancer immunology research. 2014;2(6):522-9.
22. Cho YH, Kim MS, Chung
HS, Hwang EC. Novel immunotherapy in metastatic renal cell carcinoma.
Investigative and Clinical Urology. 2017;58(4):220-7.
23. Sönmez MG, Sönmez LÖ.
New treatment modalities with vaccine therapy in renal cell carcinoma. Urology
Annals. 2019;11(2):119-25.
24. Amato RJ. Vaccine
therapy for renal cell carcinoma. Reviews in Urology. 2003;5(2):65.
25. Finn OJ. Human tumor
antigens, immunosurveillance, and cancer vaccines. Immunologic research.
2006;36:73-82.
26. Hole N, Stern P. A 72 kD
trophoblast glycoprotein defined by a monoclonal antibody. British journal of
cancer. 1988;57(3):239-46.
27. Rahma OE, Ashtar E,
Ibrahim R, Toubaji A, Gause B, Herrin VE, et al. A pilot clinical trial testing
mutant von Hippel-Lindau peptide as a novel immune therapy in metastatic renal
cell carcinoma. Journal of translational medicine. 2010;8:1-9.
28. Gerlinger M, Rowan AJ,
Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor
heterogeneity and branched evolution revealed by multiregion sequencing. New
England journal of medicine. 2012;366(10):883-92.
29. Razafinjatovo C.
Molecular Profiling of Clear Cell Renal Cell Carcimoma and Targeted Therapy
Response: University of Zurich; 2017.
30. Fang Y, Mo F, Shou J,
Wang H, Luo K, Zhang S, et al. A pan-cancer clinical study of personalized
neoantigen vaccine monotherapy in treating patients with various types of
advanced solid tumors. Clin Cancer Res. 2020;26(17):4511-20.
31. Ott PA, Hu Z, Keskin DB,
Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine
for patients with melanoma. nature. 2017;547(7662):217-21.
32. Ott PA, Hu-Lieskovan S,
Chmielowski B, Govindan R, Naing A, Bhardwaj N, et al. A phase Ib trial of
personalized neoantigen therapy plus anti-PD-1 in patients with advanced
melanoma, non-small cell lung cancer, or bladder cancer. Cell.
2020;183(2):347-62. e24.
33. Hassanipour S, Namvar G,
Fathalipour M, Salehiniya HJB. The incidence of kidney cancer in Iran: a
systematic review and meta-analysis. 2018;8(2).
34. Gottlich HC, Nabavizadeh
R, Dumbrava M, Pessoa RR, Mahmoud AM, Garg I, et al. Emerging Antibody-Drug
Conjugate Therapies and Targets for Metastatic Renal Cell Carcinoma. Kidney
Cancer. 2023;7(1):161-72.
35. Beksac AT, Paulucci DJ,
Blum KA, Yadav SS, Sfakianos JP, Badani KK. Heterogeneity in renal cell
carcinoma. Urol Oncol. 2017 Aug;35(8):507-515.
36. Paulucci DJ, Sfakianos
JP, Yadav SS, Badani KK. BAP1 is overexpressed in black compared with white
patients with Mx-M1 clear cell renal cell carcinoma: A report from the cancer
genome atlas. Urol Oncol. 2016 Jun;34(6):259.e9-259.e14.