Evaluation of beta-hemolytic, metallic green sheen,
and ONPG test properties Escherichia coli isolated from urinary tract
infections
Hossein Karamy Ghadikolae 1,
Majid Alipour 1,2*, Ramin Mofarrah 3
1 Department of
Cell and Molecular Biology, Babol Branch, Islamic Azad University, Babol, Iran
2
Comprehensive Health
Research Centre, Babol Branch, Islamic Azad University, Babol, Iran
3 Department of Dermatology, Faculty of Medicine, Sari Branch, Islamic
Azad University, Sari, Iran
*Corresponding Author: Majid Alipour
* Email: alipourmk@gmail.com
Abstract
Introduction: Uropathogenic Escherichia coli strains are the most common
cause of urinary tract infections in nosocomial and community-acquired
infections. Phenotypic characteristics of Escherichia coli isolates in patients
with urinary tract infections vary from region to region. Therefore, studying
the phenotypic properties of the bacterium is very important.
Materials and Methods: In the current study, 100 strains of Escherichia coli were
detected from urine samples of patients with urinary tract infections in
Mazandaran province, Babol. This study aimed to investigate the properties of
metallic green sheen, beta hemolysis, and
Ortho-nitrophenyl-β-D-galactopyranoside (ONPG) test of Escherichia coli.
Results: The most common bacterium isolated from urinary tract infections was E.
coli (68.02%). In the present study, the properties of beta hemolysis,
metallic green sheen, and ONPG in uropathogenic E. coli were 1, 80, and
100%, respectively.
Conclusion: The results of this study showed that 20% of E. coli strains lacked
metallic green sheen, which should be identified through the IMViC test and
other biochemical tests.
Keywords: Uropathogenic E. coli, ONPG test, Metallic green sheen, β
hemolysis
Introduction
Escherichia coli (E. coli) is normally found in the intestines of humans and
animals (1). Escherichia coli is the most common cause of UTI,
accounting for 80 to 90 percent of community-acquired infections and 40 to 50
percent of nosocomial infections. More than 50% of women between the ages of 20
and 40 experience a urinary tract infection more than once (2). If left
untreated, cystitis can accelerate ascending infections such as pyelonephritis
and sepsis with kidney damage (3). E. coli that cause urinary tract
infections are known as uropathogenic E. coli (UPEC) (4). Urinary tract
infections are caused by bacteria ascending from around the urethra to the
urethra, bladder, and other urinary tract. Colonization of the area around the
urethra with pathogenic bacteria is a vital factor in the development of
urinary tract infections. Women are more likely than men to get urinary tract
infections due to the shorter distance between the urethra and the proximity of
the urethra to the anus (5). Phenotypic tests of metallic green sheen in the
culture medium of eosin methylene blue agar, hemolysis in blood agar medium, and
Ortonitrophenyl beta galactoside are used in the detection of uropathogenic Escherichia
coli. In EMBs, strong acid producers such as E. coli usually form
green metallic-colored colonies that show a dark core. Under acidic conditions,
the color of eosin Y precipitates, and due to the formation of an amide bond
between eosin Y and methylene blue in the medium, dark-colored colonies usually
form with a metallic green glow (6). uropathogenic E.
coli lysis red blood cell by producing hemolysin A by creating a hole in
the membrane. HlyA, encoded by the hlyCABD operon, is the most important factor
in the UPEC. This toxin gives UPEC the ability to cause tissue damage, cross
mucosal barriers, release host nutrients, and damage immune cells (7). The ONPG
test detects lactose-negative and delayed lactose-positive bacteria. Prevalence
of lactose-negative strains (0-5%) has been reported in studies (8). Given that
the beta-hemolytic, metallic green sheen, and ONPG characteristics of
uropathogenic E. coli have been reported differently, this study aimed to
investigate these properties.
Materials and Methods
Sample collection and detection
In this cross-sectional descriptive
epidemiological study, urine samples were collected from patients referred to
Babol Shahid Beheshti Hospital, Mazandaran Province, Babol. A total of 1202
urine samples were collected for 5 months (from November 2020 to March 2020) under the supervision of the Medical Ethics Committee of the Islamic Azad
University, Babol branch (Ethics Code 1345). Blood agar, McConkey
agar, and Eosin methylene blue (EMB) agar media were used for isolation and identification of
UPEC. If the number of colonies of a single microorganism in the urine sample
was 105 colony-forming units per milliliter (CFU / ml) or more, a
urinary tract infection would be considered positive. Patients who received
antibiotics two weeks before sample collection were excluded from the investigation (9). Strains of E. coli were confirmed using the IMViC (Indole,
Methyl Red, Voges-Proskauer, Citrate utilization), urease and triple sugar iron
agar (TSIA), and other biochemical tests. All confirmed isolates were stored in
trypticase soy broth with 15% glycerol at -80 ° C until further investigation
(10).
Metallic green sheen
EMB (Eosin Methylene Blue) agar medium is a
selective and differential culture medium used to isolate facultative anaerobic
bacteria such as E. coli (11). To determine the green sheen, E. coli colonies are cultivated as streak plate on EMB agar
medium, then are kept in an incubator at 37◦C for 24 hours. Lactose-fermenting
Gram-negative bacteria acidify the environment and under acidic conditions the
dyes produce a dark purple complex, which is usually accompanied by a metallic
green sheen. metallic green sheen is an indicator of intense fermentation of
lactose and/or sucrose. A smaller amount of acid production resulting
from slow fermentation develops a brown-pink coloration of growth. Colonies of
non-lactose fermenters appear as clear or pink (12).
Ortho-nitrophenyl-β-D-galactopyranoside test
Lactose permease and beta-galactosidase
enzymes are needed to break down lactose. β-galactosidase permease exists in
the cytoplasmic membrane in which transports lactose into the cell, but
cytoplasmic beta-galactosidase hydrolyzes lactose into glucose and galactose.
There are strains of E. coli that ferment lactose with delay. Some bacteria
lack β-galactoside permase enzyme but have β-galactosidase (13). In these
strains, there is an insertion sequence between the genes of beta-galactosidase
and lactose permease, which may cause a decrease in the expression of lactose
permease. To identify delayed lactose fermenting bacteria, the ONPG test is
used (14). ONPG is structurally similar to lactose and colorless and enters the
cell more than lactose. Inside the cell, ONPG is cleaved by β-galactosidase to
o-nitrophenol, which has a yellow color. To perform the test, one ONPG disc is
add to a sterile test tube containing 0.1 ml of sterile 0.85% w/v sodium
chloride solution (physiological saline). Emulsify the desired colony in the
tube containing the disc. Incubate the tube at 37-35 degrees Celsius. Observe
the tube at 1- to 6 hour intervals to detect active lactose fermenters.
Incubate the tubes for 24 hours to detect late lactose fermenters. If
beta-galactosidase is positive, the fluid and disc will turn yellow (15).
Hemolysis on blood agar
Certain bacteria produce extracellular
hemolysin, which hemolyzes red blood cells and releases the hemoglobin
contained in them. three types hemolysis create on blood agar medium. Alpha hemolysis
creates a green halo around the colony. The hemolysis is produced by oxidation
of oxyhemoglobin (Fe+2 ) to
non-oxygen-binding met-hemoglobin (Fe+3) through hydrogen peroxide
(16). Beta hemolysis, the complete destruction of
red blood cells, shows a clear area around the colony. Gamma hemolysis
indicates the absence of hemolysis around the colony. Blood agar
culture medium is usually prepared from trypticase soy agar or Columbia agar
base with 5% sheep blood. Pure colonies were inoculated using streak
plate method on blood agar and then, were kept at 37°C for 24 hours.
Results
In urine culture, 147 individuals had significant urinary tract infections caused by Gram-negative bacteria. The average
age of patients was 58.2 years. The prevalence of urinary tract infections
caused by E. coli strains was 100 (68.02%). Out of 100 Escherichia
coli isolates, 80% showed a metallic green sheen but 20% did not have a
green gloss (Figure 1).
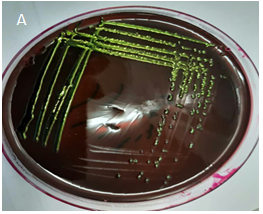

Figure 1. A. Metallic green sheen, B. No
green sheen.
The characteristic of beta hemolysis from 100
tested Escherichia coli isolates on Blood agar medium showed that only
one (1%) of them had beta hemolysis and the rest (99%) did not have hemolysis (Figure
2).
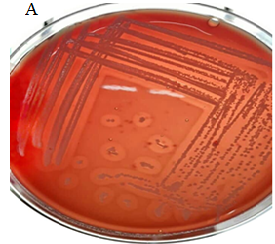
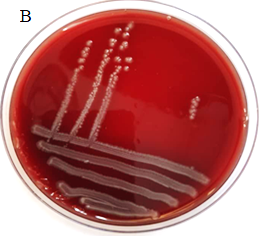
Figure 2. A. Beta hemolysis, B. Gamma
hemolysis.
The ONPG test of all E. coli strains
showed that 100% of the strains had positive reaction (Figure 3).

Figure 3. A. Positive ONPG, B. Negative ONPG
Discussion
Escherichia coli is the most prevalent bacterial agent that causes urinary tract
infections (UTIs), mainly in women. In our research, the uropathogenic E.
coli was responsible for 68.02% of urinary tract infections. In a study by
Michael W. Dunne and colleagues, it was shown that E. coli caused 75.6%
of UTIs (17). In another study, was determined that 60.5% of UTI was caused by E.
coli (18). The results of these studies show that UTIs caused by E. coli
have almost the same prevalence. In the current research, 80% of uropathogenic E.
coli showed metallic green sheen in an EMB agar medium. In the study
conducted by Jain et al., all strains of E. coli separated from the
urine had a metallic green sheen (19). The presence of metallic green sheen has
been reported in various studies, but the prevalence of green sheen caused by
uropathogenic Escherichia coli has not been determined definitively. In
our study, only one percent of uropathogenic E. coli showed beta
hemolysis, but the rest did not cause hemolysis. In the study carried out by
Sayan Bhattacharyya, only 10% of uropathogenic E. coli were hemolytic (20). In
the study conducted by Sonal Jindal, 34% of uropathogenic E. coli produced
hemolysin and 66% of the remaining isolates did not show hemolysis (21). Noha
Mahmoud showed that 15 (30%) uropathogenic E. coli isolates were
β-hemolytic while 35 isolates (70%) were non-hemolytic (22). The results of
these studies show that the production of hemolysin in uropathogenic E. coli
is different in different regions, which is a common occurrence due to
mutations in the gene. Based on our results, 100% of uropathogenic E. coli
isolates were ONPG positive. In a study by C. LONGHI and et al., 70.9% of
uropathogenic E. coli presented ONPG positive (23). In another study
conducted by Mahshid Deldar Abad Paskeh , all 91 isolates of uropathogenic E.
coli were ONPG positive (24). The results of these investigations are
almost consistent with the present study.
Conclusions
In conclusion, all uropathogenic E. coli don’t produce metallic
green sheen, so other biochemical features must be considered. A small
percentage of uropathogenic E. coli cause beta hemolysis, so this test
can be used to identify E. coli in the laboratory. Almost all uropathogenic E. coli are ONPG positive, so this test is
not mandatory.
Author contribution
MA designed research, analyzed the data, wrote the manuscript, and
performed the interpretation of the results; RM wrote the manuscript and
collected the specimens; HKGh performed the practical experiments and
collected the samples.; All authors read and approved the final manuscript.
Acknowledgments
The authors express their gratitude and appreciation to all people
who contributed to this manuscript.
Conflict of interest
The authors declare that there is no conflict of interest in this
manuscript.
References
1. El-Baz R, Said HS,
Abdelmegeed ES, Barwa R. Characterization of virulence determinants and
phylogenetic background of multiple and extensively drug-resistant Escherichia
coli isolated from different clinical sources in Egypt. Appl Microbiol
Biotechnol. 2022;106(3):1279-98.
2. Zhu H, Chen Y, Hang Y,
Luo H, Fang X, Xiao Y, et al. Impact of inappropriate empirical antibiotic
treatment on clinical outcomes of urinary tract infections caused by
Escherichia coli: a retrospective cohort study. J Glob Antimicrob Resist.
2021;26:148-53.
3. Tutone M, Johansen TEB,
Cai T, Mushtaq S, Livermore DM. Susceptibility and Resistance to Fosfomycin and
other antimicrobial agents among pathogens causing lower urinary tract
infections: findings of the SURF study. Int J Antimicrob Agents.
2022;59(5):106574.
4. Tanabe RH, Dias RC,
Orsi H, de Lira DR, Vieira MA, Dos Santos LF, et al. Characterization of
Uropathogenic Escherichia coli Reveals Hybrid Isolates of Uropathogenic and
Diarrheagenic (UPEC/DEC) E. coli. Microorganisms. 2022;10(3):645.
5. Muriuki CW, Ogonda LA,
Kyanya C, Matano D, Masakhwe C, Odoyo E, et al. Phenotypic and genotypic
characteristics of uropathogenic Escherichia coli isolates from Kenya. Microb
Drug Resist. 2022;28(1):31-8.
6. Divya P, Paul S,
Fathima P, Abdulla MH. Comparative evaluation of EMB agar and hicrome E. coli
agar for differentiation of green metallic sheen producing non E. Coli and
typical E. Coli colonies from food and environmental samples. J Pure Appl Microbiol. 2016;10(4):2863-70.
7. Derakhshan S, Ahmadi S,
Ahmadi E, Nasseri S, Aghaei A. Characterization of Escherichia coli isolated
from urinary tract infection and association between virulence expression and
antimicrobial susceptibility. BMC Microbiol. 2022;22(1):1-11.
8. Behzadi P, Urbán E,
Gajdács M. Association between biofilm-production and antibiotic resistance in
uropathogenic Escherichia coli (UPEC): an in vitro study. Diseases.
2020;8(2):17.
9. Jomehzadeh N, Saki M,
Ahmadi K, Zandi G. The prevalence of plasmid-mediated quinolone resistance
genes among Escherichia coli strains isolated from urinary tract infections in
southwest Iran. Mol Biol Rep. 2022:1-7.
10. Heidarlo MN, Lotfollahi
L, Yousefi S, Lohrasbi V, Irajian G, Talebi M. Analysis of virulence genes and
molecular typing of Listeria monocytogenes isolates from human, food, and
livestock from 2008 to 2016 in Iran. Trop Anim Health Prod. 2021;53(1):1-9.
11. Sharma P, Melkania U.
Enhancement effect of amino acids on hydrogen production from organic fraction
of municipal solid waste using co-culture of Escherichia coli and Enterobacter
aerogenes. Energy convers manag.
2018;163:260-7.
12. Lal A, Cheeptham N.
Eosin-methylene blue agar plates protocol. Am Soc Microbiol. 2007.
13. Sharma G, Dang S, Kalia
M, Gabrani R. Synergistic antibacterial and anti-biofilm activity of nisin like
bacteriocin with curcumin and cinnamaldehyde against ESBL and MBL producing
clinical strains. Biofouling. 2020;36(6):710-24.
14. Gill A, McMahon T,
Dussault F, Jinneman K, Lindsey R, Martin H, et al. Delayed lactose utilization
among Shiga toxin-producing Escherichia coli of serogroup O121. Food Microbiol.
2022;102:103903.
15. Chauhan A, Jindal T.
Biochemical and molecular methods for bacterial identification. Microbiological Methods for Environment, Food
and Pharmaceutical Analysis: Springer; 2020. p. 425-68.
16. McDevitt E, Khan F,
Scasny A, Thompson CD, Eichenbaum Z, McDaniel LS, et al. Hydrogen Peroxide
Production by Streptococcus pneumoniae Results in Alpha-hemolysis by Oxidation
of Oxy-hemoglobin to Met-hemoglobin. Msphere. 2020;5(6):e01117-20.
17. Dunne MW, Puttagunta S,
Aronin SI, Brossette S, Murray J, Gupta V. Impact of Empirical Antibiotic
Therapy on Outcomes of Outpatient Urinary Tract Infection Due to Nonsusceptible
Enterobacterales. Microbiol spectr. 2022;10(1):e02359-21.
18. Horie A, Nariai A, Katou
F, Abe Y, Saito Y, Koike D, et al. Increased community-acquired upper urinary
tract infections caused by extended-spectrum beta-lactamase-producing
Escherichia coli in children and the efficacy of flomoxef and cefmetazole. Clin
Exp Nephrol. 2019;23(11):1306-14.
19. Jain P, Bepari AK, Sen
PK, Rafe T, Imtiaz R, Hossain M, et al. High prevalence of multiple antibiotic
resistance in clinical E. coli isolates from Bangladesh and prediction of
molecular resistance determinants using WGS of an XDR isolate. Sci rep.
2021;11(1):1-13.
20. Bhattacharyya S, Sarfraz
A, Ansari MAA, Jaiswal N. Characterization and antibiogram of uropathogenic
Escherichia coli from a tertiary care hospital in Eastern India. Int J Curr
Microbiol Appl Sci. 2015;4(2):701-5.
21. Jindal S, Shivani M.
Study on phenotypic assays to determine virulence factors of uropathogenic
escherichia coli (UPEC) isolates and their correlation with antibiotic
resistance pattern in tertiary care hospital of western Uttar Pradesh. Indian J
Basic Appl Med Res.2018;7(4): 275-282.
22. Gohar NM, Aly HF, Ayoub
MI. Important Virulence Factors and Related Genes in Uropathogenic E. coli and
their Relation to Fluoroquinolone Resistance. J Pure Appl Microbiol.
2018(3):1393-403.
23. Longhi C, Cossu A, Iebba
V, Massaro M, Cipriani D, Chiarini F, et al. Virulence traits in Escherichia
coli strains isolated from outpatients with urinary tract infections. IntJ
Immunopathol and Pharmacol. 2008;21(3):715-23.
24. Paskeh MDA, Moghaddam
MJM, Salehi Z. Prevalence of plasmid-encoded carbapenemases in multi-drug
resistant Escherichia coli from patients with urinary tract infection in
northern Iran. Iran J Basic Med Sci. 2020;23(5):586.