Variable
performance of lncRNA in breast cancer
Sogand Vahidi 1*, Fatemeh Nejatifar 2*, Mostafa Khaleghipoor 3, Habib Eslami Kenarsari
4
1 Clinical Research Development Unit of Poursina Hospital, Guilan University of Medical Sciences, Rasht, Iran
2 Department of Hematology and
Oncology, Razi hospital, School of Medicine, Guilan
University of Medical Sciences, Rasht, Iran
3 Anesthesia,
Neyshabur University of Medical Sciences, Neyshabur, Iran
4 Inflammatory Lung Diseases Research Center, Department of Internal
Medicine, Razi Hospital, Guilan University of Medical
Sciences, Rasht, Iran
*Corresponding Authors:
Sogand Vahidi * Email: so.vahidii@gmail.com
Fatemeh Nejatifar * Email: dr.f.nejatifar@gmail.com
Abstract
Introduction: Breast cancer, is one of most frequent cancers across women, is
recognized as a diverse and difficult disease that continues to be a serious
public health problem. Long non-coding RNAs have already attracted a lot of
interest as a result of the advancement of next-generation sequencing methods.
Various studies indicate that long non-coding RNAs play an essential part in tumor
growth. Even though the biological purpose and molecular processes of long
non-coding RNAs are still unknown, modern data has shown that a variety of long
non-coding RNAs express inappropriately in malignancies, particularly breast
cancer. This review highlighted the most recent research on long non-coding
RNAs in breast cancer, with an emphasis on the many molecular functions of
regulatory long non-coding RNAs.
Keywords: Breast cancer, Long non-coding RNA, Cell
proliferation, Molecular mechanisms
Introduction
The cancer
susceptibility is to some extent due to the inheritance of significant genetic
factors that vary depending on the type of cancer. Breast cancer is one of the
most common causes of death in women, especially in industrialized countries (1). Breast cancer screening allows
early detection of malignancy and ultimately reduced mortality. Although a
variety of imaging techniques are commonly used to screen for breast cancer,
they often lack sufficient sensitivity and diagnostic specificity (2). Therefore, access to appropriate,
reliable, accurate, non-invasive, as well as cost-effective diagnostic methods
is needed to identify breast tissue abnormalities. ncRNAs involved in cancer
have been identified by a variety of techniques, including expression
microarrays, tiling arrays, methylation analysis and next-generation sequence (3).
Biogenesis
and roles of lncRNA in BC
One
of the most influential factors in the development of cancer that is widely
studied today is the genes that regulate cancer pathways. Recent advances in
RNA biology show that non-coding RNAs are essential molecules (4). They have specific regulatory
functions in the formation and progression of diseases, especially cancer. To
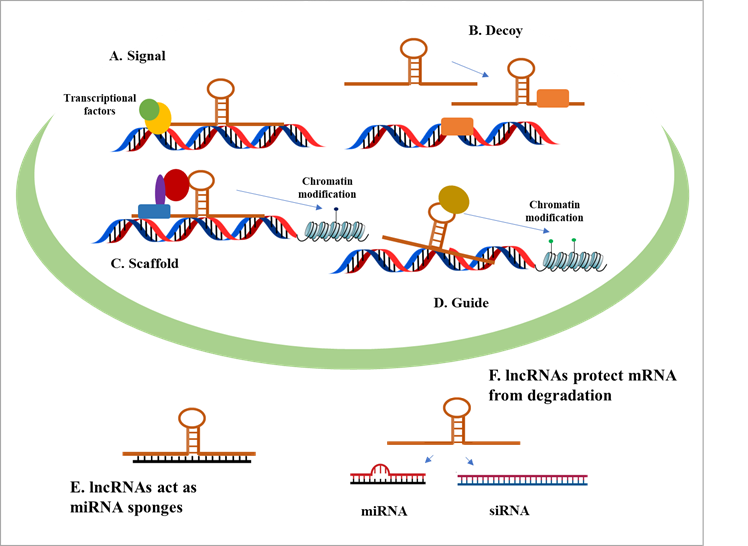
organize the processes of lncRNA activity, numerous categorization systems have
been developed. One of them categorizes lncRNAs into
four items including signal, decoy, scaffold and guide (Figure 1).

Figure 1. Roles of long noncoding RNAs (lncRNAs).
Breast cancer is the leading cause of death in women's health.
Despite improvements in gene regulation of breast cancer, and even
individualized therapies are being developed based on four molecular types
(Luminal A, Luminal B, Her2 positive, and triple-negative breast cancer
(TNBC)), but still failing to decrease the occurrence and overall death rate.
Numerous long noncoding RNAs (lncRNAs) have been
linked to breast cancer (3, 5). LncRNAs
are a kind of ncRNA transcript that regulates gene expression at the
transcriptional, translational, and post-translational stages; nevertheless, lncRNAs do not produce peptides or proteins, while being
essential for cell types to work properly. LncRNAs
perform their tasks in a variety of ways, such as interchromosomal
contact mediation, functioning as sponges for endogenous RNAs, controlling mRNA
decay, and altering epigenetic modifications that are steered to their
destinations, among others (4, 6, 7). As a result,
any variation in lncRNA expression levels can result in a variety of disorders,
notably cancer (8). There is also abundant awareness
that lncRNAs may control gene expression at the
post-transcriptional stage. MiRNAs function as post-transcriptional controls of
their messenger RNA (mRNA) targets via mRNA degradation as essential
cytoplasmic controllers. There are a vast number of LncRNAs
(9, 10). However, the transcripts of
numerous forms of LncRNAs are not stable across
specimens with comparable genetic links, and only around 200 types of LncRNAs have been studied in detail thus far. As a
consequence, researchers are wondering if all LncRNAs
have biological activities, and further study is needed to solve this topic.
The majority of lncRNAs are found in the nucleus and
chromatin, where they govern DNA sequences and are engaged in transcriptional
regulation with various activities in the cytoplasm, while a subset of
molecules is found in the cytoplasm as circulating lncRNAs,
which are conveyed by exosomes (11).
LncRNAs can interact with a wide range of molecules, comprising
transcription factors, mature mRNAs, chromatin-modifying complexes, RNA-associated
proteins, DNA, nascent RNA transcripts, microRNA, and chromatin. LncRNA
transcripts can bind to active proteins and determine their precise location (12). As a result, lncRNAs
play critical roles in regulating gene expression at the epigenetic, transcriptional,
and post-transcriptional stages. Different lncRNAs
have been identified and studied in breast cancer that is involved in various
processes of tumor formation, proliferation and cell migration. For example,
ATB, HOXA-AS2 and CCAT2 increase
expression in tumor tissues compared to margin tissues, resulting in increased
proliferation, cell migration, and metastasis (13) (Table 1).
|
lncRNAs |
Rols |
Ref |
|
NKILA |
Epithelial
mesenchymal transition |
(14) |
|
XIST |
Cell growth
and metastasis |
(15) |
|
PTENP1 |
Migration and
proliferation |
(16) |
|
ANCR |
Metastasis
and invasion |
(17) |
|
MEG3 |
Epithelial-mesenchymal transition and proliferation |
(18) |
|
PDCD4-AS1 |
Progression |
(19) |
|
MAGI2-AS3 |
Cell growth |
(20) |
|
Lnc015192 |
Epithelial-mesenchymal
transition, invasion and migration |
(21) |
|
GACAT3 |
Proliferation |
(22) |
|
CHET1 |
Invation,
proliferation and migration |
(23) |
|
TUG1 |
Apoptosis,
migration and proliferation |
(24) |
|
PVT1 |
Proliferation |
(25) |
|
ATB |
Epithelial
mesenchymal transition |
(26) |
|
NNT-AS1 |
Progression |
(27) |
|
NEAT1 |
Metastasis |
(28) |
|
UCA1 |
Metastasis |
(29) |
Retinoblastoma tumor suppressor (RB) is an important regulator of
the cell cycle and a large number of processes associated with tumor growth.
Functional inactivation of RB has been identified sporadically in many human
tumors, which is involved in the onset or progression of the disease. Numerous
studies have now shown that the disappearance of this tumor suppressor creates
a selective vulnerability that can be therapeutically targeted and thus provide
an accurate approach to exploiting RB deficiency (30, 31).
RB is believed to be
inactivated as a result of two different mechanisms in breast cancer. One of
these mechanisms is the loss of RB gene as a result of homozygous deletion in
breast cancer of triple-negative type and the second pathway is through
phosphorylation by CDK4/6 (32).
HOTAIR expression is significantly increased in breast tumors, and
measuring its levels is a defining indicator in the diagnosis of primary breast
tumors, the likelihood of metastasis occurring, and patient survival. LncRNAs are categorized into two groups based on their
involvement in the development of BC, those that stimulate the development of
BC and those that hinder the development of BC (33). The role of HOTAIR in breast cancer
metastasis has been demonstrated. By targeting the PRC2 set and directing it to
a specific gene set, it reinforces the expression patterns that promote
aggression and migration. HOTAIR expression is increased 2,000-fold in
metastatic breast cancer specimens (1, 34).
The function of H19 has also been demonstrated in the development
of metastases, including in breast cancer. SNP rs2107425 in the intron 1 of H19
gene is significantly associated with short-term survival without metastasis (35).
Whether they stimulate or hinder the formation of BC, their
mechanism of action typically encompasses the following aspects, influence BC
cell proliferation and apoptosis, influence BC cell invasion and influence BC
cell treatment resistance (36).
Several LncRNAs stimulate the growth of
BC, and their roles have been studied in the preliminary stage. It aids in the
development of more effective methods for diagnosing BC, determining its
prognosis, forecasting its origin, and interfering with therapy. Mechanisms
associated to these LncRNAs will be discussed in
detail below (37).
Up to this point, many LncRNAs that limit
BC formation have already been thoroughly investigated. They have been shown to
primarily impede BC formation by reducing proliferation or promoting apoptosis (38).
Negatively affecting BC cell migration and invasion
Researchers discovered a novel LncRNA called NF-KappaB
associating LncRNA (NKILA). It is elevated by NF-B and connects to NF-B/IB to
form stable composites that effectively cover the phosphorylated structural
regions of IB. As a result, IKK (IB kinase) triggered IB phosphorylation and
NF-B activation (39). Furthermore, NKILA may inhibit
excessive NF-B activation in mammary epithelial cells in response to
inflammatory stimuli. NKILA may increase apoptosis and decrease invasion in
MDA-MB-231 cells. To summarize, NKILA may limit BC proliferation and metastasis
via suppressing NF-B function (40).
As a result, certain lncRNAs may hinder
the genesis and maintenance of BC. In respect of function, it primarily
inhibits the genesis and growth of BC by lowering proliferation of BC cells,
encouraging apoptosis of such cells, and preventing cell invasion and
metastasis (41). Nonetheless, few LncRNAs have now been identified to be efficient for
suppressing BC, and even fewer were studied in terms of their reaction
mechanisms (42).
Conclusion
Previously assumed to be transcriptional
background, lncRNAs, like miRNAs, are now generally
recognized as key regulators of gene expression and cancer. The lncRNAs are prospective
strategies for human cancer detection, treatment, and therapy. Surprisingly,
abnormal lncRNA expression is linked to breast cancer. In contrast to
protein-coding mRNAs and miRNAs, our comprehension of lncRNAs
is still in its early stages. There is a major
gap in the understanding of lncRNAs. It's unclear if
aberrant lncRNA expression is a factor or a result of carcinogenesis. As a
result of the growing amount of lncRNAs discovered,
their biological activities and methods of action in cancer deserve additional
investigation. There are a lot of lncRNAs in the
bloodstream. Nevertheless, research into circulating lncRNAs
in cancer is still in its initial phases. A considerable
study is required before circulating lncRNAs may be
used as diagnostic, prognostic, or therapeutic indicators. Finally, the
identification of lncRNAs has led to advances in
cancer study. lncRNAs have the potential to play a
key role in cancer screening, prognosis, and treatment advancement, aiding
patients with breast cancer and others.
Author contributions
SV, FN, MKh, and HEK
wrote and compiled this article. SV wrote and edited the manuscript
comprehensively. All authors confirmed the final version of the paper.
Conflict of interest
The authors declare that they have no conflicts of interest.
References
1. Mozdarani H, Ezzatizadeh V, Parvaneh RR. The
emerging role of the long non-coding RNA HOTAIR in breast cancer development
and treatment. J Transl Med. 2020;18(1):1-15.
2. Freeman K, Geppert J, Stinton C, Todkill D,
Johnson S, Clarke A, et al. Use of artificial intelligence for image analysis
in breast cancer screening programmes: systematic review of test accuracy. BMJ.
2021;374.
3. Kim T, Reitmair A. Non-coding RNAs: functional
aspects and diagnostic utility in oncology. Int J Mol Sci. 2013;14(3):4934-68.
4. Statello L, Guo C-J, Chen L-L, Huarte M. Gene
regulation by long non-coding RNAs and its biological functions. Nat Rev Mol
Cell Biol. 2021;22(2):96-118.
5. Li C, Zhao Z, Zhou J, Liu Y, Wang H, Zhao X.
Relationship between the TERT, TNIP1 and OBFC1 genetic polymorphisms and
susceptibility to colorectal cancer in Chinese Han population. Oncotarget.
2017;8(34):56932.
6. Grillone K, Riillo C, Scionti F, Rocca R,
Tradigo G, Guzzi PH, et al. Non-coding RNAs in cancer: platforms and strategies
for investigating the genomic “dark matter”. J Exp Clin Cancer Res.
2020;39(1):1-19.
7. Dizaji BF. Strategies to target long non-coding
RNAs in cancer treatment: progress and challenges. Egypt J Med Hum Genet.
2020;21(1):1-15.
8. Arun G, Diermeier SD, Spector DL. Therapeutic
targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018;24(3):257-77.
9. Marín-Béjar O, Huarte M. Long noncoding RNAs:
from identification to functions and mechanisms. Adv Genet. 2015;5:257-74.
10. Yoon
J-H, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long
noncoding RNA. J Mol Biol. 2013;425(19):3723-30.
11. Kung JT, Colognori D, Lee JT. Long noncoding
RNAs: past, present, and future. Genetics. 2013;193(3):651-69.
12. Schmitz SU, Grote P, Herrmann BG. Mechanisms
of long noncoding RNA function in development and disease. Cell Mol Life Sci.
2016;73(13):2491-509.
13. Li Y, Yang X, Kang X, Liu S. The regulatory
roles of long noncoding RNAs in the biological behavior of pancreatic cancer. Saudi
J Gastroenterol. 2019;25(3):145.
14. Wu W, Chen F, Cui X, Yang L, Chen J, Zhao J,
et al. LncRNA NKILA suppresses TGF‐β‐induced epithelial–mesenchymal transition
by blocking NF‐κB signaling in breast cancer. Int J Cancer.
2018;143(9):2213-24.
15. Zheng R, Lin S, Guan L, Yuan H, Liu K, Liu C,
et al. Long non-coding RNA XIST inhibited breast cancer cell growth, migration,
and invasion via miR-155/CDX1 axis. Biochem Biophys Res Commun.
2018;498(4):1002-8.
16. Shi X, Tang X, Su L. Overexpression of long
noncoding RNA PTENP1 inhibits cell proliferation and migration via suppression
of miR-19b in breast cancer cells. Oncol Res. 2018;26(6):869.
17. Li Z, Hou P, Fan D, Dong M, Ma M, Li H, et al.
The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and
metastasis of breast cancer. Cell Death Differ. 2017;24(1):59-71.
18. Zhang W, Shi S, Jiang J, Li X, Lu H, Ren F.
LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421
targeting E-cadherin in breast cancer. Biomed Pharmacother. 2017;91:312-9.
19. Jadaliha M, Gholamalamdari O, Tang W, Zhang Y,
Petracovici A, Hao Q, et al. A natural antisense lncRNA controls breast cancer
progression by promoting tumor suppressor gene mRNA stability. PLoS Genet.
2018;14(11):e1007802.
20. Yang Y, Yang H, Xu M, Zhang H, Sun M, Mu P, et
al. Long non-coding RNA (lncRNA) MAGI2-AS3 inhibits breast cancer cell growth
by targeting the Fas/FasL signalling pathway. Hum Cell. 2018;31(3):232-41.
21. Huang X, Xie X, Liu P, Yang L, Chen B, Song C,
et al. Adam12 and lnc015192 act as ceRNAs in breast cancer by regulating
miR-34a. Oncogene. 2018;37(49):6316-26.
22. Zhong H, Yang J, Zhang B, Wang X, Pei L, Zhang
L, et al. LncRNA GACAT3 predicts poor prognosis and promotes cell proliferation
in breast cancer through regulation of miR-497/CCND2. Cancer Biomark.
2018;22(4):787-97.
23. Song R, Zhang J, Huang J, Hai T. Long
non-coding RNA GHET1 promotes human breast cancer cell proliferation, invasion
and migration via affecting epithelial mesenchymal transition. Cancer Biomark.
2018;22(3):565-73.
24. Fan S, Yang Z, Ke Z, Huang K, Liu N, Fang X,
et al. Downregulation of the long non-coding RNA TUG1 is associated with cell
proliferation, migration, and invasion in breast cancer. Biomed Pharmacother.
2017;95:1636-43.
25. Tang J, Li Y, Sang Y, Yu B, Lv D, Zhang W, et
al. LncRNA PVT1 regulates triple-negative breast cancer through
KLF5/beta-catenin signaling. Oncogene. 2018;37(34):4723-34.
26. Li R-H, Chen M, Liu J, Shao C-C, Guo C-P, Wei
X-L, et al. Long noncoding RNA ATB promotes the epithelial− mesenchymal
transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis
in breast cancer. Cell Death Dis. 2018;9(12):1-16.
27. Li Y, Lv M, Song Z, Lou Z, Wang R, Zhuang M.
Long non-coding RNA NNT-AS1 affects progression of breast cancer through
miR-142-3p/ZEB1 axis. Biomed Pharmacother. 2018;103:939-46.
28. Li W, Zhang Z, Liu X, Cheng X, Zhang Y, Han X,
et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of
hormonally responsive breast cancer. J Clin Invest. 2017;127(9):3421-40.
29. Li G-Y, Wang W, Sun J-Y, Xin B, Zhang X, Wang
T, et al. Long non-coding RNAs AC026904. 1 and UCA1: a “one-two punch” for
TGF-β-induced SNAI2 activation and epithelial-mesenchymal transition in breast
cancer. Theranostics. 2018;8(10):2846.
30. Vélez-Cruz R, Johnson DG. The retinoblastoma
(RB) tumor suppressor: pushing back against genome instability on multiple
fronts. Int J Mol Sci. 2017;18(8):1776.
31. Wu T, Wu L. The Role and Clinical Implications
of the Retinoblastoma (RB)-E2F Pathway in Gastric Cancer. Front Oncol.
2021;11:1954.
32. Rocca A, Farolfi A, Bravaccini S, Schirone A,
Amadori D. Palbociclib (PD 0332991): targeting the cell cycle machinery in
breast cancer. Expert Opin Pharmacother. 2014;15(3):407-20.
33. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X,
Brugmann SA, et al. Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311-23.
34. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM,
Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to
promote cancer metastasis. Nature. 2010;464(7291):1071-6.
35. Riaz M, Berns EM, Sieuwerts AM,
Ruigrok-Ritstier K, de Weerd V, Groenewoud A, et al. Correlation of breast
cancer susceptibility loci with patient characteristics, metastasis-free
survival, and mRNA expression of the nearest genes. Breast Cancer Res Treat. 2012;133(3):843-51.
36. Wang M, Zhang C, Song Y, Wang Z, Wang Y, Luo
F, et al. Mechanism of immune evasion in breast cancer. Onco Targets Ther.
2017;10:1561.
37. Sun Z, Liu J, Liu J. The expression of
lncRNA-MALAT1 in breast cancer patients and its influences on prognosis. Cell
Mol Biol. 2020;66(3):72-8.
38. Naz F, Tariq I, Ali S, Somaida A, Preis E,
Bakowsky U. The Role of Long Non-Coding RNAs (lncRNAs) in Female Oriented
Cancers. Cancers. 2021;13(23):6102.
39. Zhao S, Zhang X, Chen S, Zhang S. Long
noncoding RNAs: fine-tuners hidden in the cancer signaling network. Cell Death
Discov. 2021;7(1):1-10.
40. Ni M-J, Hu Z-H, Liu Q, Liu M-F, Lu M-h, Zhang
J-S, et al. Identification and characterization of a novel non-coding RNA
involved in sperm maturation. PLoS One.
2011;6(10):e26053.
41. Bin X, Hongjian Y, Xiping Z, Bo C, Shifeng Y,
Binbin T. Research progresses in roles of LncRNA and its relationships with
breast cancer. Cancer Cell Int. 2018;18(1):1-12.
42. Chandra Gupta S, Nandan Tripathi Y. Potential
of long non‐coding RNAs in cancer patients: From biomarkers to therapeutic
targets. Int J Cancer. 2017;140(9):1955-67.