Comprehensive
RNA-seq analysis of alternative splicing events that distinguishes between
metastatic oral cancer of gingiva and tongue
Vishwas Sharma 1 †, Dinesh Kumar 2
†, Harpreet Singh 3, Sanjay Gupta 1 *
1 Division of Cytopathology,
ICMR-National Institute of Cancer Prevention and Research, Noida, Uttar
Pradesh, India
2 ICMR-National Institute of Cancer
Prevention and Research, Noida, Uttar Pradesh, India
3 Informatics, Systems
& Research Management (ISRM), ICMR, New Delhi
*Corresponding
Author: Sanjay Gupta * Email: sanjaydr17@gmail.com
† Equal contributiom
Abstract
Introduction: Oral cancer (OC) is a multifactorial disease caused due to various
genomic changes. Alternative splicing (AS) is a regulatory genetic process
through which messenger RNA forms diverse protein variants. This study aims to
study the variation in the AS events at tongue and gingiva locations of OC.
Materials
and Methods: Forty-five paired end OC RNA-seq data were downloaded from Sequence
Read Archive (SRA) data repository. Twenty four paired end OC (tongue 13,
gingival 11) RNA sequences passed the stringent inclusion/ exclusion criteria
which were analyzed following Tuxedo pipeline. The ClueGO (v2.5.8) tool in
Cytoscape app manager (v3.7.1) was used for gene set enrichment analysis
keeping false discovery rate (FDR <=0.05).
Results: Eighty-three genes were identified to be significantly alternatively
spliced when comparison was made between RNA sequences from normal tissues and
tumor tissues from the gingiva region (p<0.05). Similarly, 39 genes were
found to be significantly alternatively spliced when comparison was made
between normal tissues and tumor tissues from tongue region of OC. Of these,
only 4 genes i.e. AHR, AL356488.2, KREMEN1, SH3TC1
were similar in gingiva and tongue whereas others were unique to their location.
Conclusion: Genome-wide AS events vary considerably in gingival and tongue locations
of OC. Hence, these events need to be thoroughly investigated for defining the
treatment strategy. Further functional studies are needed to decipher the role
of AS in OC.
Keywords: Gingiva cancer, Oral cancer, RNA-seq, Alternative splicing, Tongue
cancer
Introduction
Oral cancer (OC) is the sixth most common type of cancer in
the world. As per the Globocan 2020 data, the global new cases diagnosed with
this disease accounted for ~3.7 million, of which ~1.7 million died (1). OC
encompasses cancer occurring at various locations such as lips, tongue,
gingiva, cheeks, the floor of the mouth, hard and soft palate, sinuses, and
throat. Oral carcinogenesis is a multi-step process that involves the
accumulation of various genomic alterations leading to aberrations in the genetic
landscape (2). Knowledge of these variations may help decide the therapeutic
strategy for treatment.
The alternative splicing (AS) or differential splicing is a
regulatory genetic mechanism of
choosing different combinations of splice-sites in an mRNA precursor (pre-mRNA)
to form variably spliced mRNAs. These events contribute significantly to the
etiopathogenesis of OC. In our previous systematic review, we have highlighted
the RNA-seq based studies which depicted the role of AS in Head and Neck cancer
(HNC) (3). Through this review, we have noticed that the role of AS events at
different locations of OC not been addressed in the available
literature. To fill this research gap, we aim to study the variability in the
AS events at two different locations of OC i.e. tongue and gingiva.
Materials and Methods
In order to understand the AS events
at different locations of OC, the screening of the RNA-seq data was undertaken
on Sequence Read Archive (SRA) data repository on 29th September 2020. Briefly,
the search terms “oral cancer [All Fields] AND "Homo sapiens"[orgn]
AND ("biomol rna"[Properties] AND "library layout
paired"[Properties] AND "platform illumina"[Properties] AND
"filetype fastq"[Properties])” were used for screening the
data/sequences. The detailed search strategy is illustrated in Figure 1.
Briefly, a total of 45 paired-end RNA-seq data were obtained from SRA. Of
these, 21 paired-end sequences were on OC cell lines and hence excluded from
the study (Table 1). The remaining 24 paired-end OC (tongue 13, gingival 11)
sequences were included following the inclusion/exclusion criteria mentioned
below. All these sequences were from a study by Bhattacharya et al. (4).
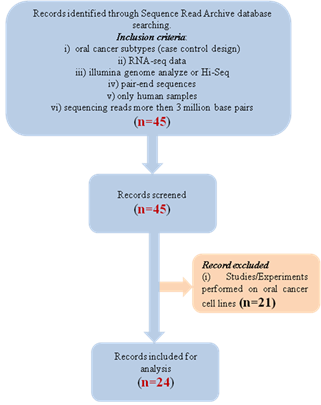
Figure
1. Detailed description of the records being screened.
Inclusion criteria
i) Oral cancer at the selected
location (case-control design).
ii) RNA-seq data.
iii) Sequencing performed on
Illumina genome analyzer or Hi-Seq.
iv) Pair-end sequencing reads.
Exclusion criteria
i) Sequencing performed on a
platform other than Illumina.
ii) Samples other than humans.
iii) Sequencing reads less than 3
million base pairs.
The raw fastq reads were downloaded
and accessed for high-quality sequences using the FastQC program. The software
Trimmomatic (v0.40) was used for trimming the sequences and removing the
adapter content. For trimming, the parameters HEADCROP:12 TRAILING:1 SLIDINGWINDOW:4:20
MINLEN:50 were used. The mapping of the reads with the human genome (GRCh38)
was done through Tophat using default parameters, followed by assembly through
Cufflinks. The differentially spliced genes were identified through Cuffdiff
using Jensen–Shannon divergence test between isoforms. P-value <0.05 was
considered to be significant. The ClueGO (v2.5.8) tool in Cytoscape app manager
(v3.7.1) was used for gene set enrichment analysis (GSEA) keeping false
discovery rate (FDR <=0.05), two-sided hypergeometric test with Bonferroni
step down correction and kappa score 0.4 with other default parameter was used
for GO biological process and molecular function.
Table
1. Details of screening and samples being excluded or included.
|
Sample type
|
Layout
|
Inclusion/Exclusion remark
|
|
RNAseq_HSC2_10mM
|
Paired end
|
Oral cancer
cell line HSC2 treated with 10mM metformin. Since the study is on cell line
it is excluded from the analysis.
|
|
RNAseq_HSC2_control
|
Paired end
|
Oral
cancer cell line HSC2 without Metformin. Since the study is on cell line it
is excluded from the analysis.
|
|
RNAseq_Cal27_10mM
|
Paired end
|
Oral cancer cell line Cal27 treated with 10mM metformin. Since
the study is on cell line it is excluded from the analysis.
|
|
RNAseq_Cal27_control
|
Paired
end
|
Oral
cancer cell line Cal27 without Metformin. Since the study is on cell line it
is excluded from the analysis.
|
|
GSM4726150: tongue9-A64034; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726149: tongue8-A64033; Homo sapiens; RNA-Seq
|
Paired
end
|
Included
|
|
GSM4726148: tongue7-A64032; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726147: tongue6-A64031; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726146: tongue5-A64029;
Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726145: tongue4-A64028; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726144: tongue3-A64027; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726143: tongue2-A64025; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726142: tongue10-A64036; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726141: tongue1-A64024; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726140: tcont3-A64037; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726139: tcont2-A64035; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726139: tcont2-A64035; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726137: gingiva9-A64048; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726136: gingiva8-A64047; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726135: gingiva7-A64046; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726134: gingiva6-A64045; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726133: gingiva5-A64044; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726132: gingiva4-A64043; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726131: gingiva3-A64041; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726130: gingiva2-A64040; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726129: gingiva1-A64039; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726128: gcont2-A64042; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4726127:
gcont1-A64038; Homo sapiens; RNA-Seq
|
Paired end
|
Included
|
|
GSM4550990:
TW2.6; Homo sapiens; RNA-Seq
|
Paired end
|
Oral cancer
cell line TW2. Since the study is on cell line it is excluded from the
analysis
|
|
GSM4550989: OEC-M1; Homo sapiens; RNA-Seq
|
Paired end
|
Oral
cancer cell line OEC-M1. Since the study is on cell line it is excluded from
the analysis
|
|
GSM4550988: OC3; Homo sapiens; RNA-Seq
|
Paired end
|
Oral cancer
cell line OC3. Since the study is on cell line it is excluded from the
analysis
|
|
GSM4550987: CGHNC9; Homo sapiens; RNA-Seq
|
Paired end
|
Oral
cancer cell line CGHNC9. Since the study is on cell line it is excluded from
the analysis
|
|
GSM4550986: CGHNC8; Homo sapiens; RNA-Seq
|
Paired end
|
Oral cancer cell line CGHNC9. Since the study is on cell line it
is excluded from the analysis
|
|
GSM4550985: CGHNK6; Homo sapiens; RNA-Seq
|
Paired end
|
Oral
Keratinocyte cell line CGHNK6. Since the study is on cell line it is excluded
from the analysis
|
|
GSM4550984: CGHNK2; Homo sapiens; RNA-Seq
|
Paired end
|
Oral Keratinocyte cell line CGHNK2. Since the study is on cell
line it is excluded from the analysis
|
|
RNA-Seq_SAS_control
|
Paired end
|
Human
tongue squamous cell carcinoma cell line control. Since the study is on cell
line it is excluded from the analysis
|
|
RNA-Seq_SAS_HGK
|
Paired end
|
Human tongue squamous cell carcinoma cell line case treated with
Hydroxygenkwanin drug. Since the study is on cell line it is excluded from
the analysis
|
|
RNA-Seq_OECM_control
|
Paired end
|
Human
gingival squamous carcinoma cell line control. Since the study is on cell
line it is excluded from the analysis
|
|
RNA-Seq_OECM_HGK
|
Paired end
|
Human gingival squamous carcinoma cell line treated with
Hydroxygenkwanin drug. Since the study is on cell line it is excluded from
the analysis
|
|
HuR-CP1 (HuR cleavage product-1)+134; Homo sapiens; RNA-Seq
|
Paired end
|
Oral
cancer cell line transfected with gene HuR-CP1 (HuR cleavage product-1).
Since the study is on cell line it is excluded from the analysis
|
|
HuR-D226A (asp-aln mutant)_133; Homo sapiens; RNA-Seq
|
Paired end
|
Oral cancer cell line transfected with gene HuR-D226A (asp-aln
mutant)_133. Since the study is on cell line it is excluded from the analysis
|
|
HUR
FL (full-length HuR)_132; Homo sapiens; RNA-Seq
|
Paired end
|
Oral
cancer cell line transfected with gene HUR FL (full-length HuR)_132. Since
the study is on cell line it is excluded from the analysis
|
|
GFP control_131; Homo sapiens; RNA-Seq
|
Paired end
|
Oral cancer
cell line control (GFP control). Since the study is on cell line it is
excluded from the analysis
|
|
CELF1 KD replicate 1; Homo sapiens; RNA-Seq
|
Paired end
|
Si
CELF1 treated tongue suqamous cell carcinoma cell line. Since the study is on
cell line it is excluded from the analysis
|
|
WT replicate 1; Homo sapiens; RNA-Seq
|
Paired end
|
Tongue squamous cell carcinoma cell line control. Since the
study is on cell line it is excluded from the analysis
|
Results
A total of 83 genes were found to be significantly
alternatively spliced when a comparison was made between normal tissues and
tumor tissue samples obtained from the gingiva region (p<0.05). Similarly,
39 genes were found to be significantly alternatively spliced when a comparison
was made between normal tissues and tumor tissue samples obtained from the
tongue region (Table 2).
Of these, only 4 genes
i.e. AHR, AL356488.2, KREMEN1, SH3TC1 were found to
be commonly alternatively spliced in gingiva as well as tongue whereas others
were unique to the location, that is, either gingiva or tongue (Figure 2).
Table 2. Details of differentially spliced
genes in samples obtained from gingiva and tongue region of oral cancer.
|
Gingiva
|
|
Tongue
|
|
AC010323.1, NDUFA7
|
|
ABR
|
|
AHR
|
|
AHR
|
|
AL049629.2
|
|
AKR1C2
|
|
AL356488.2
|
|
AL356488.2
|
|
ANKRD11
|
|
AP1S1
|
|
ASCC2
|
|
ATP6V1E1
|
|
ASDURF, ASNSD1
|
|
ATP9B
|
|
ATF5
|
|
ATXN1
|
|
ATP13A2
|
|
BAZ1B
|
|
ATP5F1B
|
|
CDC37, MIR1181
|
|
BID
|
|
COL4A5
|
|
CCT5
|
|
COL5A1
|
|
COL12A1
|
|
DIS3L2
|
|
COL7A1
|
|
EGLN1
|
|
CORO2A
|
|
FBLIM1
|
|
CPSF1
|
|
GAMT
|
|
CTU2
|
|
GNB1
|
|
E2F3
|
|
IFI16
|
|
EMP3
|
|
JUP
|
|
ERAP1
|
|
KREMEN1
|
|
EXOC4
|
|
LIMK2
|
|
FBLN2
|
|
LMF2
|
|
FEZ1
|
|
MED14
|
|
FMNL2
|
|
NAPRT
|
|
GFOD1
|
|
NFATC3
|
|
GPD2
|
|
NRBF2
|
|
GTF3C1
|
|
NSUN5
|
|
HJURP
|
|
PIGP
|
|
HMBS
|
|
PLEKHG5
|
|
HP1BP3
|
|
S100A13
|
|
HSPBP1
|
|
SH3TC1
|
|
IK,MIR3655
|
|
SLC38A7
|
|
INF2
|
|
SPP1
|
|
INPP4A
|
|
TAB3
|
|
ISOC2
|
|
TENM4
|
|
KCTD15
|
|
TRIB2
|
|
KIAA1217
|
|
TYK2
|
|
KREMEN1
|
|
USP22
|
|
LMTK2
|
|
VWA1
|
|
LPXN
|
|
|
|
MARF1
|
|
|
|
MCM3AP
|
|
|
|
MGAT4B
|
|
|
|
NDRG1
|
|
|
|
NMD3
|
|
|
|
NOL7
|
|
|
|
PAFAH1B1
|
|
|
|
PDCD11
|
|
|
|
PIP5K1C
|
|
|
|
PLCD3
|
|
|
|
PNKP
|
|
|
|
POLA1
|
|
|
|
PPP1CC
|
|
|
|
PPP1R14B-AS1
|
|
|
|
PRRC2A
|
|
|
|
PSMD2
|
|
|
|
SACM1L
|
|
|
|
SEC14L1
|
|
|
|
SH3TC1
|
|
|
|
SLC3A2
|
|
|
|
SNIP1
|
|
|
|
STARD3NL
|
|
|
|
SUN1
|
|
|
|
TAF13
|
|
|
|
TASP1
|
|
|
|
TBC1D23
|
|
|
|
TDRD7
|
|
|
|
TGFA
|
|
|
|
TJP2
|
|
|
|
TMEM131L
|
|
|
|
TMEM138
|
|
|
|
TMEM177
|
|
|
|
TNFRSF10A
|
|
|
|
TNRC6B
|
|
|
|
TPM1
|
|
|
|
TTC38
|
|
|
|
TXNIP
|
|
|
|
UGP2
|
|
|
|
VPS18
|
|
|
|
YBX3
|
|
|
|
ZC3HAV1
|
|
|
|
ZFAND6
|
|
|
|
ZMIZ1
|
|
|
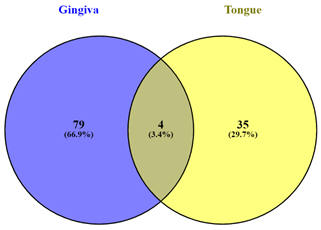
Figure 2. Details of differentially spliced
genes in samples obtained from gingiva and tongue region of oral cancer.
Gene set enrichment analysis
Gingiva: The GSEA of AS genes revealed their
significant association with cell death cycles, vesicle targeting, and
tethering processes, inositol phosphate metabolism process, Charcot-Marie-Tooth
disease, and cerebral cortex radial glia guided migration signaling (Figure 3a).
Tongue: The GSEA of the genes AS in the tongue
revealed their role in prostaglandin signaling, formation of collagen trimers,
poly-pyrimidine binding, and regulation of neuron projection regeneration (Figure
3b).
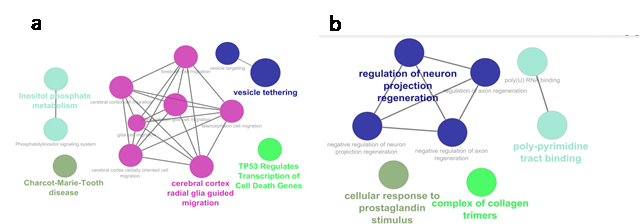
Figure 3. Depiction of gene set enrichment
analysis for biological processes and cellular signals associated with differentially
spliced genes in tongue (a) and gingiva (b).
Discussion
The result of this
study suggests that variability in the AS events exists at the location level
in OC. Hence, to understand the etiology of OC at the genetic level, the
location from where the samples are obtained is an important parameter that
needs to be considered. Understanding the signature of AS events, at the gene
level, at different locations of OC may help us in developing molecular
target/s for early prediction/diagnosis of OC. Moreover, it may also be useful
in deciding the best therapeutic option from the available ones, for the
management/treatment of OC.
The gene set
enrichment analysis (GSEA) of the alternatively spliced genes revealed various
genes whose role in cancer development has already been well defined, for
instance, the genes that code for INPP4A, and PLCD3 were found to be
alternatively spliced, play a significant role in inositol phosphate metabolism.
The suppressed INPP4A gene leads to
enhanced PIP3 level which promotes AKT1-dependent tumor growth and subsequent
metastasis (5). The phospholipase C delta 3, i.e. PLCD3 is anti apoptosis
molecule that plays a significant role in cancer cell proliferation and
migration. The other gene CCT5 chaperonin has been found to be upregulated in
p53-mutated tumors (6) and has been suggested as a potential cancer biomarker
(7). The ZC3HAV1 regulates KRAS and acts as metastasis-promoting factor in
pancreatic cancer (8). Another gene EXOC4
and VPS18 play an imperative role in
vesicles docking involved in exocytosis. The gene prostaglandin E2 (PGE2) acts as a stimulator for tumor
progression (9). High expression of the prostaglandin receptors has been observed
in squamous cell carcinomas. The genes coding for AKR1C2 and GNB1 proteins were
found to be differentially spliced which are linked with prostaglandin
stimulus. We also noticed differential splicing in mRNA of genes coding for
COL4A5 and COL5A1 in tongue and COL12A1 and COL7A1 in gingival cancerous
tissue, which is responsible for the formation of collagen trimers and the
formation of larger collagen molecules. Finally higher expression of Ataxin-1
(ATXN1), a proto-oncogene, has been reported in cervical cancer and considered
as a potent tumor genetic factor (10). Cancer-causing genes show differential
splicing which projects the roles of different isoforms for tumorigenesis and
maintaining the mRNA expression to a threshold tumorigenic stage. Overall, the
genes related to carcinogenesis, cancer progression, and metastasis show AS
which points toward the role of different isoforms to establish the cancer
cells.
The functional
validation of the AS genes is needed to confirm the findings. However, we report
the genes whose role in other cancers is already well defined. Further, studies
at other locations of OC such as lips, floor of the mouth, etc are needed to decipher
the role of AS events in metastatic OC. We were unable to segregate the data
based on nodal status in metastatic OC as directed in the study by Bhattacharya
et el.,4. Studying AS in OC in relation to the nodal
status will be helpful to understand the association between its genetic and
clinical features. We have performed analysis on 24 paired-end OC samples that
were freely available in the data repositories. However, due to the limited
sample size, there is a possibility that some genes might be alternatively
spliced but have been missed as they did not reach the significant statistical
cutoff. A more profound analysis of a
larger dataset is required to critically analyze and understand the role of AS
events in OC.
Conclusion
Genome-wide AS
events at different locations of OC vary which needs thorough investigation for
defining the treatment strategy.
Author
contributions
VS and SG designed the study, VS and DK
collected the information and analyzed the data, and wrote the first draft of
the manuscript. HS reviewed the manuscript. SG led the group, reviewed,
and wrote the final draft.
Conflict of interest
The authors declare that they have no competing interests.
Funding
This work was funded by the Indian Council of Medical Research by
grant No. 2019-3557, this supported the fellowship of Dr. Vishwas Sharma.
Acknowledgments
The authors are thankful to Dr. Amit Kumar for providing critical
suggestions on the manuscript. The grammar was checked by Grammarly software
and the plagiarism was checked by iThenticate software.
References
1. Globocan 2020 fact
sheethttps://gco.iarc.fr/today/data/factsheets/cancers/1-Lip-oral-cavity-fact-sheet.pdf
(2020).
2. Ram H, Sarkar J, Kumar H, et al. Oral Cancer: Risk Factors and
Molecular Pathogenesis. J Maxillofac Oral Surg. 2011; 10: 132–137.
3. Sharma V, Nandan A, Singh H, et al. Events of alternative
splicing in head and neck cancer via RNA sequencing - an update. BMC Genomics.
2019; 20: 442.
4. Bhattacharya A, Janal MN, Veeramachaneni R, et al. Oncogenes
overexpressed in metastatic oral cancers from patients with pain: potential
pain mediators released in exosomes. Sci Rep. 2020; 10: 14724.
5. Ye Y, Jin L, Wilmott JS, et al. PI(4,5)P2 5-phosphatase A
regulates PI3K/Akt signalling and has a tumour suppressive role in human
melanoma. Nat Commun. 2013; 4: 1508.
6. Ooe A, Kato K, Noguchi S. Possible involvement of CCT5, RGS3, and
YKT6 genes up-regulated in p53-mutated tumors in resistance to docetaxel in
human breast cancers. Breast Cancer Res Treat. 2007; 101: 305–315.
7. Gao H, Zheng M, Sun S, et al. Chaperonin containing TCP1 subunit
5 is a tumor associated antigen of non-small cell lung cancer. Oncotarget.
2017; 8: 64170–64179.
8. Huang W, Hua H, Xiao G, et al. ZC3HAV1 promotes the proliferation
and metastasis via regulating KRAS in pancreatic cancer. Aging (Albany NY).
2021; 13: 18482–18497.
9. Hoshikawa. Expression of prostaglandin E2 receptors in oral
squamous cell carcinomas and growth inhibitory effects of an EP3 selective
antagonist, ONO-AE3-240. Int J Oncol. 2009; 34.
10. Kang A-R, An H-T, Ko J, et al. Ataxin-1 regulates
epithelial-mesenchymal transition of cervical cancer cells. Oncotarget. 2017;
8: 18248–18259.


